

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146548.2 + phase: 0
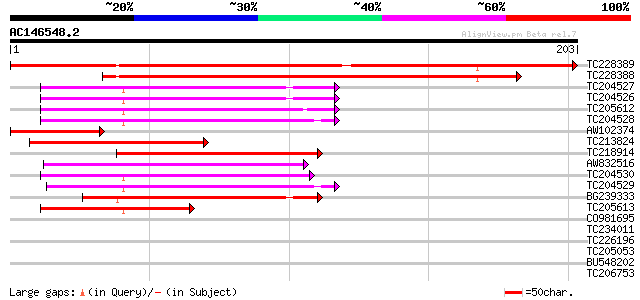
(203 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC228389 weakly similar to UP|Q7X857 (Q7X857) Amelogenin-like pr... 313 3e-86
TC228388 weakly similar to UP|Q7X857 (Q7X857) Amelogenin-like pr... 204 3e-53
TC204527 70 7e-13
TC204526 70 7e-13
TC205612 similar to GB|AAM91395.1|22137100|AY133565 At4g14410/dl... 70 7e-13
TC204528 66 9e-12
AW102374 64 5e-11
TC213824 similar to UP|Q9LT23 (Q9LT23) Emb|CAA18500.1, partial (... 62 2e-10
TC218914 similar to UP|Q9LT23 (Q9LT23) Emb|CAA18500.1, partial (... 61 4e-10
AW832516 61 4e-10
TC204530 55 2e-08
TC204529 51 3e-07
BG239333 47 5e-06
TC205613 45 2e-05
CO981695 38 0.004
TC234011 37 0.006
TC226196 UP|Q8L5W2 (Q8L5W2) BZIP transcription factor ATB2, comp... 35 0.023
TC205053 homologue to UP|Q8L5W2 (Q8L5W2) BZIP transcription fact... 35 0.023
BU548202 35 0.030
TC206753 weakly similar to UP|GUN4_ARATH (Q9LX31) Tetrapyrrole-b... 34 0.040
>TC228389 weakly similar to UP|Q7X857 (Q7X857) Amelogenin-like protein,
partial (22%)
Length = 919
Score = 313 bits (803), Expect = 3e-86
Identities = 161/205 (78%), Positives = 179/205 (86%), Gaps = 2/205 (0%)
Frame = +1
Query: 1 MKQGKVPKRIHKAEREKMKREHLNELFLDLANALDLSEPNNGKASILIEASRLLKDLLCQ 60
M QGKVP++IHKAEREKMKREHLNE F+DLA+ALDL+E NNGKASIL E +RLLKDLL Q
Sbjct: 247 MNQGKVPRKIHKAEREKMKREHLNERFVDLASALDLNE-NNGKASILCETARLLKDLLSQ 423
Query: 61 IQSLKKENVSLLSESHYVTMEKNELKEENSSLETQIEKLQGEIQARIAQSKPDLNAPPQL 120
I+SLKKENV+LLSESHY+TMEKNELKEEN SLETQIEKLQGEIQAR+AQSKPDLN PP
Sbjct: 424 IESLKKENVTLLSESHYMTMEKNELKEENCSLETQIEKLQGEIQARLAQSKPDLNVPPH- 600
Query: 121 ELESPEQTTFSGQSFQLPTVDPTLQQGPTVLVVPFRPDLQAAYPAP--TEITPNPPLVIS 178
E PEQT F GQ+ QL T++P LQQG TVLVVPF P+LQA++PAP TE+TP V+S
Sbjct: 601 --EPPEQTNFPGQNLQLHTIEPNLQQGSTVLVVPFHPNLQASFPAPNVTEVTPKSMSVVS 774
Query: 179 KPHARYPTPADSWPSQLLGEQPTSS 203
KPHARYPTPADSWPSQLLGEQPTSS
Sbjct: 775 KPHARYPTPADSWPSQLLGEQPTSS 849
>TC228388 weakly similar to UP|Q7X857 (Q7X857) Amelogenin-like protein,
partial (21%)
Length = 454
Score = 204 bits (518), Expect = 3e-53
Identities = 107/152 (70%), Positives = 123/152 (80%), Gaps = 2/152 (1%)
Frame = +1
Query: 34 LDLSEPNNGKASILIEASRLLKDLLCQIQSLKKENVSLLSESHYVTMEKNELKEENSSLE 93
LDL+E NNGKASIL E +RLLKDLL QI+SLKKENV+LLSES+Y+TMEKNELKEEN SLE
Sbjct: 1 LDLNE-NNGKASILCETARLLKDLLSQIESLKKENVTLLSESNYMTMEKNELKEENCSLE 177
Query: 94 TQIEKLQGEIQARIAQSKPDLNAPPQLELESPEQTTFSGQSFQLPTVDPTLQQGPTVLVV 153
TQIEKLQG+IQAR+AQ KPDLN PP LELE EQT F GQ+ QL T++P LQQG +LVV
Sbjct: 178 TQIEKLQGQIQARLAQCKPDLNVPPHLELEPLEQTNFPGQNLQLHTIEPNLQQGSAILVV 357
Query: 154 PFRPDLQAAYPAP--TEITPNPPLVISKPHAR 183
P PDL ++PAP T + P V+SKPHAR
Sbjct: 358 PCNPDLPPSFPAPNVTGVMPKSTSVVSKPHAR 453
>TC204527
Length = 898
Score = 70.1 bits (170), Expect = 7e-13
Identities = 41/108 (37%), Positives = 64/108 (58%), Gaps = 1/108 (0%)
Frame = +2
Query: 12 KAEREKMKREHLNELFLDLANALDLSEP-NNGKASILIEASRLLKDLLCQIQSLKKENVS 70
KA REK++R+ LN+ F++L + L+ P KASILI+A+R++ L + LK N S
Sbjct: 428 KACREKLRRDRLNDKFVELGSILEPGRPPKTDKASILIDAARMVTQLRDEALKLKDSNTS 607
Query: 71 LLSESHYVTMEKNELKEENSSLETQIEKLQGEIQARIAQSKPDLNAPP 118
L + + EKNEL++E L+ + EKL E+Q + ++P PP
Sbjct: 608 LQEKIKELKAEKNELRDEKQRLKAEKEKL--EVQVKSMNAQPAFLPPP 745
>TC204526
Length = 807
Score = 70.1 bits (170), Expect = 7e-13
Identities = 41/108 (37%), Positives = 64/108 (58%), Gaps = 1/108 (0%)
Frame = +2
Query: 12 KAEREKMKREHLNELFLDLANALDLSEP-NNGKASILIEASRLLKDLLCQIQSLKKENVS 70
KA REK++R+ LN+ F++L + L+ P KASILI+A+R++ L + LK N S
Sbjct: 41 KACREKLRRDRLNDKFVELGSILEPGRPPKTDKASILIDAARMVTQLRDEALKLKDSNTS 220
Query: 71 LLSESHYVTMEKNELKEENSSLETQIEKLQGEIQARIAQSKPDLNAPP 118
L + + EKNEL++E L+ + EKL E+Q + ++P PP
Sbjct: 221 LQEKIKELKAEKNELRDEKQRLKAEKEKL--EVQVKSMNAQPAFLPPP 358
>TC205612 similar to GB|AAM91395.1|22137100|AY133565 At4g14410/dl3245w
{Arabidopsis thaliana;} , partial (47%)
Length = 863
Score = 70.1 bits (170), Expect = 7e-13
Identities = 43/108 (39%), Positives = 63/108 (57%), Gaps = 1/108 (0%)
Frame = +3
Query: 12 KAEREKMKREHLNELFLDLANALDLSEP-NNGKASILIEASRLLKDLLCQIQSLKKENVS 70
KA REK++RE LNE F DL++ L+ P K +IL +A R+L L + Q LKK N
Sbjct: 174 KACREKLRRERLNERFCDLSSVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKKTNEK 353
Query: 71 LLSESHYVTMEKNELKEENSSLETQIEKLQGEIQARIAQSKPDLNAPP 118
LL E + EKNEL+EE L+ E+++ +++A + + APP
Sbjct: 354 LLEEIKCLKAEKNELREEKLVLKADKERIEKQLKA-LPVAPAGFMAPP 494
>TC204528
Length = 793
Score = 66.2 bits (160), Expect = 9e-12
Identities = 40/108 (37%), Positives = 63/108 (58%), Gaps = 1/108 (0%)
Frame = +2
Query: 12 KAEREKMKREHLNELFLDLANALDLSEP-NNGKASILIEASRLLKDLLCQIQSLKKENVS 70
KA REK++R+ LN+ F++L L+ P KA+ILI+A R++ L + Q LK N
Sbjct: 368 KACREKLRRDRLNDKFVELGAILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDTNQG 547
Query: 71 LLSESHYVTMEKNELKEENSSLETQIEKLQGEIQARIAQSKPDLNAPP 118
L + + EKNEL++E L+ + EKL+ ++++ AQ P PP
Sbjct: 548 LQEKIKELKAEKNELRDEKQRLKAEKEKLEQQLKSLNAQ--PSFMPPP 685
>AW102374
Length = 393
Score = 63.9 bits (154), Expect = 5e-11
Identities = 29/34 (85%), Positives = 33/34 (96%)
Frame = +2
Query: 1 MKQGKVPKRIHKAEREKMKREHLNELFLDLANAL 34
M QGKVP++IHKAEREKMKREHLN+LFLDLA+AL
Sbjct: 140 MNQGKVPRKIHKAEREKMKREHLNDLFLDLASAL 241
>TC213824 similar to UP|Q9LT23 (Q9LT23) Emb|CAA18500.1, partial (18%)
Length = 544
Score = 61.6 bits (148), Expect = 2e-10
Identities = 29/64 (45%), Positives = 43/64 (66%)
Frame = +3
Query: 8 KRIHKAEREKMKREHLNELFLDLANALDLSEPNNGKASILIEASRLLKDLLCQIQSLKKE 67
++ KA+REK++R+ LNE F++L N LD P N KA+I+ + +LLKDL Q+ LK E
Sbjct: 345 RKTQKADREKLRRDRLNEQFVELGNILDPDRPKNDKATIIGDTIQLLKDLTSQVSKLKDE 524
Query: 68 NVSL 71
+L
Sbjct: 525 YATL 536
>TC218914 similar to UP|Q9LT23 (Q9LT23) Emb|CAA18500.1, partial (23%)
Length = 1030
Score = 60.8 bits (146), Expect = 4e-10
Identities = 30/74 (40%), Positives = 46/74 (61%)
Frame = +2
Query: 39 PNNGKASILIEASRLLKDLLCQIQSLKKENVSLLSESHYVTMEKNELKEENSSLETQIEK 98
P N KA+IL + +LLKDL+ Q+ LK E L ES +T+EK +L+EE +SL++ I+
Sbjct: 41 PKNDKATILCDTIQLLKDLISQVSKLKDEYAMLNEESRELTLEKTDLREEKASLKSDIDN 220
Query: 99 LQGEIQARIAQSKP 112
L + Q ++ P
Sbjct: 221 LNNQYQQQLRTMFP 262
>AW832516
Length = 304
Score = 60.8 bits (146), Expect = 4e-10
Identities = 34/95 (35%), Positives = 53/95 (55%)
Frame = +2
Query: 13 AEREKMKREHLNELFLDLANALDLSEPNNGKASILIEASRLLKDLLCQIQSLKKENVSLL 72
A+R+K+ R+ LNE F ++ NALD P N KA+IL E ++L D + LK E SL
Sbjct: 2 ADRKKLTRDRLNEHFQEMGNALDPDRPKNDKATILTETVQMLIDCTADVYRLKTEYNSLA 181
Query: 73 SESHYVTMEKNELKEENSSLETQIEKLQGEIQARI 107
E + + N+L EN+S + +L + + R+
Sbjct: 182 EE*LELVPDTNDLYRENTSRHICVYRLTVQYRLRV 286
>TC204530
Length = 957
Score = 55.1 bits (131), Expect = 2e-08
Identities = 33/99 (33%), Positives = 57/99 (57%), Gaps = 1/99 (1%)
Frame = +1
Query: 12 KAEREKMKREHLNELFLDLANALDLSEP-NNGKASILIEASRLLKDLLCQIQSLKKENVS 70
KA REK++R+ LN+ F++L + L+ P KA+ILI+A ++ +K N
Sbjct: 487 KACREKLRRDRLNDKFVELGSILEPGRPAKTDKAAILIDAGQMGNQFTG*SPEVKDTNQG 666
Query: 71 LLSESHYVTMEKNELKEENSSLETQIEKLQGEIQARIAQ 109
L + + EKNEL++E L+ + EKL+ ++++ AQ
Sbjct: 667 LQEKIKELKAEKNELRDERHRLKAENEKLEQQLRSVNAQ 783
>TC204529
Length = 854
Score = 51.2 bits (121), Expect = 3e-07
Identities = 32/106 (30%), Positives = 56/106 (52%), Gaps = 1/106 (0%)
Frame = +1
Query: 14 EREKMKREHLNELFLDLANALDLSEP-NNGKASILIEASRLLKDLLCQIQSLKKENVSLL 72
+ + +K + F++L + L+ P K +ILI+A R++ L + Q LK N L
Sbjct: 34 DSDGLKESGSKKRFVELGSILEPGRPAKTDKTAILIDAVRMVTQLRGEAQKLKDTNQGLQ 213
Query: 73 SESHYVTMEKNELKEENSSLETQIEKLQGEIQARIAQSKPDLNAPP 118
+ + EKNEL++E L+ + EKL+ ++++ AQ P PP
Sbjct: 214 EKIKELKAEKNELRDEKQRLKAEKEKLEQQLKSLNAQ--PSFMPPP 345
>BG239333
Length = 376
Score = 47.4 bits (111), Expect = 5e-06
Identities = 30/87 (34%), Positives = 55/87 (62%), Gaps = 1/87 (1%)
Frame = +2
Query: 27 FLDLANALDLS-EPNNGKASILIEASRLLKDLLCQIQSLKKENVSLLSESHYVTMEKNEL 85
FL+L++ L+ S +P + K +IL +A+R++ L + + LK+ N L ++ + EKNEL
Sbjct: 44 FLELSSILEPSRQPKSDKVAILSDAARVVIQLRNEAKRLKEMNDELQAKVKELKGEKNEL 223
Query: 86 KEENSSLETQIEKLQGEIQARIAQSKP 112
++E + L+ + EKL E Q ++A +P
Sbjct: 224 RDEKNRLKEEKEKL--EQQVKVANIQP 298
>TC205613
Length = 918
Score = 45.4 bits (106), Expect = 2e-05
Identities = 26/56 (46%), Positives = 35/56 (62%), Gaps = 1/56 (1%)
Frame = +3
Query: 12 KAEREKMKREHLNELFLDLANALDLSEP-NNGKASILIEASRLLKDLLCQIQSLKK 66
KA REK++RE LNE F DL++ L+ P K +IL +A R+L L + Q LKK
Sbjct: 750 KACREKLRRERLNERFCDLSSVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKK 917
>CO981695
Length = 415
Score = 37.7 bits (86), Expect = 0.004
Identities = 18/63 (28%), Positives = 37/63 (58%)
Frame = -1
Query: 43 KASILIEASRLLKDLLCQIQSLKKENVSLLSESHYVTMEKNELKEENSSLETQIEKLQGE 102
K+ ILI+ R++ L + Q LK N L + + EKNE++++ ++ + EKL+ +
Sbjct: 358 KSXILIDDVRMVSQLRGEAQKLKDXNQGLQEKIRDLKXEKNEVRDQKQRIKAEKEKLEQQ 179
Query: 103 IQA 105
+++
Sbjct: 178 LKS 170
>TC234011
Length = 473
Score = 37.0 bits (84), Expect = 0.006
Identities = 19/47 (40%), Positives = 33/47 (69%), Gaps = 1/47 (2%)
Frame = +2
Query: 12 KAEREKMKREHLNELFLDLANALDLS-EPNNGKASILIEASRLLKDL 57
KA REK++R+ LNE FL+L++ L+ +P K ++L +A+R++ L
Sbjct: 314 KACREKLRRDKLNERFLELSSILEPGRQPKTDKVALLSDAARVVIQL 454
>TC226196 UP|Q8L5W2 (Q8L5W2) BZIP transcription factor ATB2, complete
Length = 1518
Score = 35.0 bits (79), Expect = 0.023
Identities = 16/52 (30%), Positives = 30/52 (56%)
Frame = +2
Query: 54 LKDLLCQIQSLKKENVSLLSESHYVTMEKNELKEENSSLETQIEKLQGEIQA 105
L DL+ Q+ L+KEN +L+ + T + ++ ENS L Q+ +L +++
Sbjct: 899 LDDLVSQVAQLRKENQQILTSVNITTQQYLSVEAENSVLRAQVGELSHRLES 1054
>TC205053 homologue to UP|Q8L5W2 (Q8L5W2) BZIP transcription factor ATB2,
partial (89%)
Length = 1432
Score = 35.0 bits (79), Expect = 0.023
Identities = 16/52 (30%), Positives = 30/52 (56%)
Frame = +2
Query: 54 LKDLLCQIQSLKKENVSLLSESHYVTMEKNELKEENSSLETQIEKLQGEIQA 105
L DL+ Q+ L+KEN +L+ + T + ++ ENS L Q+ +L +++
Sbjct: 872 LDDLVSQVAQLRKENQQILTSVNITTQQYLSVEAENSVLRAQVGELSHRLES 1027
>BU548202
Length = 576
Score = 34.7 bits (78), Expect = 0.030
Identities = 16/47 (34%), Positives = 28/47 (59%)
Frame = -3
Query: 60 QIQSLKKENVSLLSESHYVTMEKNELKEENSSLETQIEKLQGEIQAR 106
++++LKKE V L + + E +LKEE S L +EK++ E + +
Sbjct: 508 ELENLKKETVQLKEKDEKASKEIKQLKEELSCLSKSLEKIKSESEEK 368
>TC206753 weakly similar to UP|GUN4_ARATH (Q9LX31) Tetrapyrrole-binding
protein, chloroplast precursor (Genomes uncoulped 4),
partial (57%)
Length = 1080
Score = 34.3 bits (77), Expect = 0.040
Identities = 28/105 (26%), Positives = 42/105 (39%), Gaps = 3/105 (2%)
Frame = +3
Query: 91 SLETQIEKLQGEIQARIAQSKPD-LNAPPQLELESPEQTTFSGQSFQLPTVDPTLQQGPT 149
SL KL+ ++ I S + +P + L++P T + P PT PT
Sbjct: 33 SLILSCSKLKWQLTLSITPSNTTTIPSPNAITLQTPHPTHHFSSNTLHPLYTPT-SLSPT 209
Query: 150 VLVV--PFRPDLQAAYPAPTEITPNPPLVISKPHARYPTPADSWP 192
+L P L+ P P P PPL ++ PT +WP
Sbjct: 210 LLQTHTSLTPSLKPPLPPPP---PQPPLPLTSSATTSPTKTSNWP 335
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.128 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,038,272
Number of Sequences: 63676
Number of extensions: 112687
Number of successful extensions: 1082
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 942
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1042
length of query: 203
length of database: 12,639,632
effective HSP length: 93
effective length of query: 110
effective length of database: 6,717,764
effective search space: 738954040
effective search space used: 738954040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146548.2


