

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
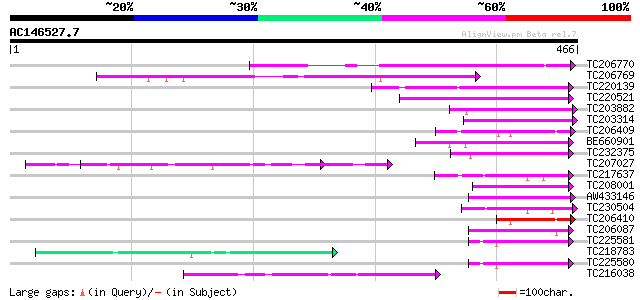
Query= AC146527.7 - phase: 0
(466 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206770 similar to UP|Q93ZG9 (Q93ZG9) AT4g25340/T30C3_20, parti... 174 1e-43
TC206769 similar to UP|Q93ZG9 (Q93ZG9) AT4g25340/T30C3_20, parti... 103 2e-22
TC220139 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl... 81 1e-15
TC220521 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl... 75 8e-14
TC203882 similar to UP|FKB2_VICFA (Q41649) FK506-binding protein... 74 1e-13
TC203314 homologue to UP|FKB2_VICFA (Q41649) FK506-binding prote... 73 2e-13
TC206409 similar to UP|Q6Z4E9 (Q6Z4E9) Immunophilin / FKBP-type ... 72 4e-13
BE660901 similar to GP|10177347|db peptidylprolyl isomerase {Ara... 68 8e-12
TC232375 similar to UP|Q9FJL3 (Q9FJL3) Peptidylprolyl isomerase,... 67 2e-11
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 67 2e-11
TC217637 similar to GB|CAD35362.1|21535744|ATH490171 FK506 bindi... 61 1e-09
TC208001 similar to UP|Q84LW7 (Q84LW7) Peptidylprolyl cis-trans ... 60 3e-09
AW433146 similar to SP|Q41649|FKB2 FK506-binding protein 2 precu... 58 1e-08
TC230504 similar to UP|Q9SCY3 (Q9SCY3) FKBP like protein , part... 58 1e-08
TC206410 similar to UP|Q6Z4E9 (Q6Z4E9) Immunophilin / FKBP-type ... 57 2e-08
TC206087 similar to UP|Q38931 (Q38931) Rof1, partial (50%) 55 5e-08
TC225581 homologue to UP|O04287 (O04287) Immunophilin, complete 55 7e-08
TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%) 54 1e-07
TC225580 homologue to UP|O04287 (O04287) Immunophilin, complete 54 2e-07
TC216038 similar to UP|Q9IZF4 (Q9IZF4) Aphid transmission P5, pa... 52 7e-07
>TC206770 similar to UP|Q93ZG9 (Q93ZG9) AT4g25340/T30C3_20, partial (23%)
Length = 929
Score = 174 bits (440), Expect = 1e-43
Identities = 101/269 (37%), Positives = 143/269 (52%), Gaps = 1/269 (0%)
Frame = +2
Query: 198 AAQDEPKMNMDEDLLTGNEQNQQQPDDKKAETTDKTLPSSQVGQGQDEKPKRKRKERSKE 257
A +D K D NE+ +PD+K AE + S V +DE
Sbjct: 2 ALKDHIKGGCDHASDNSNEKKVTEPDEKHAEDVKTSTKLSDVSHAKDE------------ 145
Query: 258 KTLFAADDACISNVVNLPQGNEHSNQDTVNGSVDNGAQDNPDGNQSEDKKVKKK-KKKSK 316
G + SN + + +KK+KKK KKK+K
Sbjct: 146 ------------------DGGKLSNNEVL-----------------VEKKIKKKNKKKTK 220
Query: 317 SQGSEVVNSDVPVSAEQSSEMMKEDGNNVEDTKPSQVRTLSNGLVIQELETGKENGKIAA 376
E + + +AE+ + ++ G +TKPSQVRT NGL+I+E+ GK +GK AA
Sbjct: 221 ESEGEAAANQITTTAEKQNLSTEKKGKKQTETKPSQVRTFPNGLIIEEVFMGKPDGKKAA 400
Query: 377 IGKKISINYTGKLKENGVVVESNAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVP 436
GKK+S+ Y GKL+++G + +SN G APFKFRLG G+VI+GW++G+ GMR+G+KRR+ +P
Sbjct: 401 PGKKVSVKYIGKLQKDGKIFDSNVGRAPFKFRLGVGQVIKGWEVGINGMRIGDKRRITIP 580
Query: 437 PSMTSKKDGESENIPPNSWLVYDFELVKV 465
PSM D +IPPNSWLV+D ELV V
Sbjct: 581 PSM-GYADKRVGSIPPNSWLVFDVELVDV 664
>TC206769 similar to UP|Q93ZG9 (Q93ZG9) AT4g25340/T30C3_20, partial (10%)
Length = 1057
Score = 103 bits (256), Expect = 2e-22
Identities = 90/339 (26%), Positives = 155/339 (45%), Gaps = 23/339 (6%)
Frame = +3
Query: 72 GKEKETKSSSNGDSTELDNAEQDEPKMNMAQDLLDQEIVDN---VDKEAVDDGK-KDGED 127
G++ E S + ++ D+ ++ D+ V N V +E VDD K ++G D
Sbjct: 117 GEDIEGTESEESSEYDSEDGYADDFIVDSDTDMYPSSPVPNSGVVIEEIVDDDKPENGYD 296
Query: 128 -----RENITIETKLKTDN------VVADIQTHR-EAEPSDQLAVPSTVPDVGDMKKSKK 175
++ + + DN +VA T E+E D +P+ V +K++
Sbjct: 297 PTKKLKKKKQVAQLKEKDNKSSGLPIVAKGDTDLVESEDEDGFPIPTAEKGVSVSQKAEA 476
Query: 176 KKKGKEKETKSSSNEHSTDLDNAAQDEPKMNMDEDLLTGNEQNQQQPDDKKAETTDKTLP 235
+ KG++ K+ + D+D++A + +K +T D+ P
Sbjct: 477 ETKGEQARKKAEKAKKEKDVDHSASVK----------------------RKVDTADEDEP 590
Query: 236 SSQVGQGQDEKPKRKRKERSKEKTLFAADDACISNVVNLPQGNEHSNQDTVNGSVDNGAQ 295
G+ +K + K KE K ++ A ++ + + + + + T+N S + A+
Sbjct: 591 QD----GKKKKKRNKLKEHIKGESDHATGNSNETKITEPDEKHPEEIKTTINLSDVSHAK 758
Query: 296 DNPDGNQS------EDKKVKKKKKKSKSQGSEVVNSDVPVSAE-QSSEMMKEDGNNVEDT 348
D DG S E K KKKKKK+K EV + + ++ E Q+ ++ G +T
Sbjct: 759 DEDDGKLSNNEVLAEKKNKKKKKKKTKESEGEVAANQITITPEKQNLSTSEKKGQKQTET 938
Query: 349 KPSQVRTLSNGLVIQELETGKENGKIAAIGKKISINYTG 387
KPSQVRT NGL+I+E+ GK +GK AA GKK+S+ Y G
Sbjct: 939 KPSQVRTFPNGLIIEEVFMGKPDGKKAAPGKKVSVKYIG 1055
>TC220139 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl isomerase
(Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase) , partial (29%)
Length = 722
Score = 80.9 bits (198), Expect = 1e-15
Identities = 49/167 (29%), Positives = 88/167 (52%), Gaps = 1/167 (0%)
Frame = +1
Query: 298 PDGNQSEDKKVKKKKKKSKSQGSEVVNSDVPVSAEQSSEMMKEDGNNVEDTKPSQVRTL- 356
P + S++ K K K S S+ S S+ + E+ ++ + ++ + K + R +
Sbjct: 16 PLSHNSKEGKTKTKTPLSFSESS----SERKAAMEEDFDIPGGEEMDLGEDKVGEEREIG 183
Query: 357 SNGLVIQELETGKENGKIAAIGKKISINYTGKLKENGVVVESNAGEAPFKFRLGKGEVIE 416
S GL + L+ G + + +G ++ ++YTG L + S ++PF F LG+G+VI+
Sbjct: 184 SRGLKKKLLKEG-QGWETPEVGDEVQVHYTGTLLDGTKFDSSRDRDSPFSFTLGQGQVIK 360
Query: 417 GWDIGLEGMRVGEKRRLVVPPSMTSKKDGESENIPPNSWLVYDFELV 463
GWD G++ M+ GE +PP + + G IPPN+ L +D EL+
Sbjct: 361 GWDEGIKTMKKGENAIFTIPPELAYGESGSPPTIPPNATLQFDVELL 501
>TC220521 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl isomerase
(Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase) , partial (40%)
Length = 890
Score = 74.7 bits (182), Expect = 8e-14
Identities = 41/143 (28%), Positives = 70/143 (48%)
Frame = +1
Query: 321 EVVNSDVPVSAEQSSEMMKEDGNNVEDTKPSQVRTLSNGLVIQELETGKENGKIAAIGKK 380
E + D + + EM +E+ K + + + + ++L E G +
Sbjct: 145 EAMGEDFEFPSAGNMEMPEEEEFESPILKVGEEKEIGKMGLKKKLLKEGEGWDTPDSGDQ 324
Query: 381 ISINYTGKLKENGVVVESNAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVPPSMT 440
+ ++YTG L + S PFKF+LG+G+VI+GWD G++ M+ GE +PP +
Sbjct: 325 VEVHYTGTLLDGTKFDSSRDRGTPFKFKLGQGQVIKGWDEGIKTMKKGENALFTIPPELA 504
Query: 441 SKKDGESENIPPNSWLVYDFELV 463
+ G IPPN+ L +D EL+
Sbjct: 505 YGESGSPPTIPPNATLQFDVELL 573
>TC203882 similar to UP|FKB2_VICFA (Q41649) FK506-binding protein 2 precursor
(Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase) (15 kDa FKBP) (FKBP-15) , partial (85%)
Length = 944
Score = 73.9 bits (180), Expect = 1e-13
Identities = 40/110 (36%), Positives = 62/110 (56%), Gaps = 5/110 (4%)
Frame = +2
Query: 362 IQELETGKENGKI-----AAIGKKISINYTGKLKENGVVVESNAGEAPFKFRLGKGEVIE 416
+ EL+ G ++ + A G ++ ++Y GKL + V S P +F LG G+VI+
Sbjct: 197 VTELQIGVKHKPVSCEVQAHKGDRVKVHYRGKLTDGTVFDSSFERNNPIEFELGTGQVIK 376
Query: 417 GWDIGLEGMRVGEKRRLVVPPSMTSKKDGESENIPPNSWLVYDFELVKVH 466
GWD GL GM +GEKR+L +P + + G IP + L++D ELV V+
Sbjct: 377 GWDQGLLGMCLGEKRKLKIPSKLGYGEQGSPPTIPGGATLIFDAELVGVN 526
>TC203314 homologue to UP|FKB2_VICFA (Q41649) FK506-binding protein 2
precursor (Peptidyl-prolyl cis-trans isomerase)
(PPIase) (Rotamase) (15 kDa FKBP) (FKBP-15) , partial
(85%)
Length = 746
Score = 73.2 bits (178), Expect = 2e-13
Identities = 37/93 (39%), Positives = 54/93 (57%)
Frame = +2
Query: 374 IAAIGKKISINYTGKLKENGVVVESNAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRL 433
+A G ++ ++Y GKL + V S P +F LG G+VI+GWD GL GM +GEKR+L
Sbjct: 185 LAHKGDRVKVHYRGKLTDGTVFDSSFERNNPIEFELGTGQVIKGWDQGLLGMCLGEKRKL 364
Query: 434 VVPPSMTSKKDGESENIPPNSWLVYDFELVKVH 466
+P + G IP + L++D ELV V+
Sbjct: 365 KIPAKLGYGDQGSPPTIPGGATLIFDTELVGVN 463
>TC206409 similar to UP|Q6Z4E9 (Q6Z4E9) Immunophilin / FKBP-type
peptidyl-prolyl cis-trans isomerase-like, partial (57%)
Length = 983
Score = 72.4 bits (176), Expect = 4e-13
Identities = 45/122 (36%), Positives = 66/122 (53%), Gaps = 7/122 (5%)
Frame = +1
Query: 351 SQVRTLSNGLVIQELETGKENGKIAAIGKKISINYTGKLKENGVVVESNA----GEAPFK 406
S TL NGL +L+ G NG A G +++I+Y K K + G P+
Sbjct: 352 SDYTTLPNGLKYYDLKVG--NGAEAKKGSRVAIHYVAKWKSITFMTSRQGMGVGGGTPYG 525
Query: 407 FRLG---KGEVIEGWDIGLEGMRVGEKRRLVVPPSMTSKKDGESENIPPNSWLVYDFELV 463
F +G +G V++G D+G++GMRVG +R L+VPP + G E IPPNS + D EL+
Sbjct: 526 FDVGQSERGTVLKGLDLGVQGMRVGGQRLLIVPPELAYGSKGVQE-IPPNSTIELDIELL 702
Query: 464 KV 465
+
Sbjct: 703 SI 708
>BE660901 similar to GP|10177347|db peptidylprolyl isomerase {Arabidopsis
thaliana}, partial (34%)
Length = 724
Score = 68.2 bits (165), Expect = 8e-12
Identities = 45/143 (31%), Positives = 68/143 (47%), Gaps = 13/143 (9%)
Frame = +2
Query: 334 SSEMMKEDGNNVEDTKPSQVRTLSNGL--VIQELETGKENGK-----------IAAIGKK 380
++E M ED + T + SN + V +E E GKE K +G +
Sbjct: 2 TAEAMGEDFDFAGATGGDLDGSSSNPILKVGEEKEIGKEGLKKKLLKEGEGWDTPEVGDE 181
Query: 381 ISINYTGKLKENGVVVESNAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVPPSMT 440
+ ++YTG L + S PF F LG+G+VI+GWD G+ M+ GE +P +
Sbjct: 182 VQVHYTGTLLDGTKFDSSRDRGTPFSFTLGQGQVIKGWDQGIITMKKGENSLFTIPAELA 361
Query: 441 SKKDGESENIPPNSWLVYDFELV 463
+ G IPPN+ L +D EL+
Sbjct: 362 YGETGSPPTIPPNATLQFDVELL 430
>TC232375 similar to UP|Q9FJL3 (Q9FJL3) Peptidylprolyl isomerase, partial
(21%)
Length = 559
Score = 67.0 bits (162), Expect = 2e-11
Identities = 37/112 (33%), Positives = 55/112 (49%), Gaps = 11/112 (9%)
Frame = +3
Query: 363 QELETGKENGKIAAI-----------GKKISINYTGKLKENGVVVESNAGEAPFKFRLGK 411
+E E GKE K + G ++ ++YTG L + S PF F LG+
Sbjct: 213 EEKEIGKEGLKKKLLKEGEGWDTPEAGDEVQVHYTGTLLDGTKFDSSRDRGTPFSFTLGQ 392
Query: 412 GEVIEGWDIGLEGMRVGEKRRLVVPPSMTSKKDGESENIPPNSWLVYDFELV 463
G+VI+GWD G+ M+ GE +P + + G IPPN+ L +D EL+
Sbjct: 393 GQVIKGWDQGIITMKKGENALFTIPAELAYGESGSPPTIPPNATLQFDVELL 548
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 67.0 bits (162), Expect = 2e-11
Identities = 68/267 (25%), Positives = 115/267 (42%), Gaps = 11/267 (4%)
Frame = +3
Query: 59 DVGDIKKSKKKKKGKEKETKSSSNGDSTEL--------DNAEQDEPKMNMAQDLLDQEIV 110
D G+ KK K KK+ K+KE K GD T++ D + +E K ++ D++
Sbjct: 6 DDGEEKKEKDKKEKKKKENKDK--GDKTDVEKVKGKENDGEDDEEKKEKKKKEKKDKDGK 179
Query: 111 DNVDK--EAVDDGKKDGEDRENITIETKLKTDNVVADIQTHREAEPSDQLAVPSTVPDVG 168
+ +K + DDGK D E +E K K D + A +E + D+ + G
Sbjct: 180 TDAEKIKKKEDDGKDDEEKKEK-----KKKYDKIDAXKVKGKEDDGKDEGNKEKKDKEKG 344
Query: 169 DMK-KSKKKKKGKEKETKSSSNEHSTDLDNAAQDEPKMNMDEDLLTGNEQNQQQPDDKKA 227
D + KK+KK KEKE K + + D +++ K + ED +E N+++ DKK
Sbjct: 345 DGDGEEKKEKKDKEKEKKKEKKDKDEETDTL-KEKGKNDEGED----DEGNKKKKKDKKE 509
Query: 228 ETTDKTLPSSQVGQGQDEKPKRKRKERSKEKTLFAADDACISNVVNLPQGNEHSNQDTVN 287
+ D + EK ++ E+ K + D I + + +
Sbjct: 510 KEKDH----------KKEKKDKEEGEKEDSKVEVSVRDIDIEEIKKEGEKEDKGK----- 644
Query: 288 GSVDNGAQDNPDGNQSEDKKVKKKKKK 314
+G ++ + + EDK K+KKKK
Sbjct: 645 ----DGGKEVKEKKKKEDKDKKEKKKK 713
Score = 55.5 bits (132), Expect = 5e-08
Identities = 52/251 (20%), Positives = 106/251 (41%), Gaps = 5/251 (1%)
Frame = +3
Query: 14 KKDSEDHENSIIETKLKTDNAFTESQIHSREAGPSDHLVDPCTVPDVGDIKKSKKKKKGK 73
KK+ + E E K K D E ++ +E D D +K KK+KK K
Sbjct: 21 KKEKDKKEKKKKENKDKGDKTDVE-KVKGKENDGED---------DEEKKEKKKKEKKDK 170
Query: 74 EKETKSSSNGDSTELDNAEQDEPKMNMAQDLLDQEIVDNVDKEAVDDGKKDGEDRENITI 133
+ +T + + ++++ + D +D V + + D+G K+ +D+E
Sbjct: 171 DGKTDAEKIKKKEDDGKDDEEKKEKKKKYDKIDAXKVKGKEDDGKDEGNKEKKDKEKGDG 350
Query: 134 ETKLKTDNVVADIQTHREAEPSDQLAVPSTVP-----DVGDMKKSKKKKKGKEKETKSSS 188
+ + K + D + ++ E D+ T+ D G+ + KKKK +KE +
Sbjct: 351 DGEEKKEK--KDKEKEKKKEKKDKDEETDTLKEKGKNDEGEDDEGNKKKKKDKKEKEKDH 524
Query: 189 NEHSTDLDNAAQDEPKMNMDEDLLTGNEQNQQQPDDKKAETTDKTLPSSQVGQGQDEKPK 248
+ D + +++ K+ + + E ++ + K + K + + + +D+K K
Sbjct: 525 KKEKKDKEEGEKEDSKVEVSVRDIDIEEIKKEGEKEDKGKDGGKEVKEKKKKEDKDKKEK 704
Query: 249 RKRKERSKEKT 259
K+K K+KT
Sbjct: 705 -KKKVTGKDKT 734
>TC217637 similar to GB|CAD35362.1|21535744|ATH490171 FK506 binding protein 1
{Arabidopsis thaliana;} , partial (60%)
Length = 870
Score = 60.8 bits (146), Expect = 1e-09
Identities = 47/126 (37%), Positives = 64/126 (50%), Gaps = 12/126 (9%)
Frame = +1
Query: 350 PSQVRTLSNGLVIQELETGKENGKIAAIGKKISINYTGKLKENGVVVESNAGEA-PFKFR 408
P + + +GL + G G A G+ I +Y G+L ENG V +S+ P FR
Sbjct: 283 PCEFQVAPSGLAFCDKLVGA--GPQAVKGQLIKAHYVGRL-ENGKVFDSSYNRGKPLTFR 453
Query: 409 LGKGEVIEGWDIGLEG------MRVGEKRRLVVPP-----SMTSKKDGESENIPPNSWLV 457
+G GEVI+GWD G+ G M G KR L +PP S + G S IPP+S L+
Sbjct: 454 VGVGEVIKGWDEGIIGGDGVPPMLAGGKRTLKIPPELGYGSRGAGCRGGSCIIPPDSVLL 633
Query: 458 YDFELV 463
+D E V
Sbjct: 634 FDVEFV 651
>TC208001 similar to UP|Q84LW7 (Q84LW7) Peptidylprolyl cis-trans isomerase ,
partial (97%)
Length = 857
Score = 59.7 bits (143), Expect = 3e-09
Identities = 28/84 (33%), Positives = 48/84 (56%), Gaps = 1/84 (1%)
Frame = +1
Query: 381 ISINYTGKLKENGVVVES-NAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVPPSM 439
+ ++Y G L + G V ++ + F F +GKG VI+ W+I ++ M+VGE ++ P
Sbjct: 253 VDVHYEGTLADTGEVFDTTHEDNTIFSFEIGKGSVIKAWEIAVKTMKVGEVAKITCKPEY 432
Query: 440 TSKKDGESENIPPNSWLVYDFELV 463
G +IPP++ LV++ ELV
Sbjct: 433 AYGSAGSPPDIPPDATLVFEVELV 504
>AW433146 similar to SP|Q41649|FKB2 FK506-binding protein 2 precursor (EC
5.2.1.8) (Peptidyl-prolyl cis- trans isomerase)
(PPiase), partial (79%)
Length = 362
Score = 57.8 bits (138), Expect = 1e-08
Identities = 31/88 (35%), Positives = 48/88 (54%)
Frame = +3
Query: 378 GKKISINYTGKLKENGVVVESNAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVPP 437
G ++ ++Y GKL + V S P +F LG G+VI+GWD L M +GEKR+L++P
Sbjct: 99 GDRVKVHYRGKLADGTVFNSSFERYYPIEFDLGTGQVIKGWDQ*LLRMSLGEKRKLIIPS 278
Query: 438 SMTSKKDGESENIPPNSWLVYDFELVKV 465
+ +G I ++ L+ LV V
Sbjct: 279 KLGCGDNGSPPTIRSSAALICGARLVVV 362
>TC230504 similar to UP|Q9SCY3 (Q9SCY3) FKBP like protein , partial (68%)
Length = 988
Score = 57.8 bits (138), Expect = 1e-08
Identities = 37/107 (34%), Positives = 60/107 (55%), Gaps = 12/107 (11%)
Frame = +1
Query: 372 GKIAAIGKKISINYTGKLKENGVVVESNAGEA-PFKFRLGKGEVIEGWDIGLEG------ 424
G A +G+ I+++YT + + G+V +S+ A P R+G G+VI+G D G+ G
Sbjct: 301 GDEAPLGELINVHYTARFAD-GIVFDSSYKRARPLTMRIGVGKVIKGLDQGILGAEGXPP 477
Query: 425 MRVGEKRRLVVPPSMTSKKD-----GESENIPPNSWLVYDFELVKVH 466
MR+G KR+L +PP + + NIP N+ L+YD V+V+
Sbjct: 478 MRIGGKRQLQIPPHLAYGPEPAGCFSGDCNIPANATLLYDINFVEVY 618
>TC206410 similar to UP|Q6Z4E9 (Q6Z4E9) Immunophilin / FKBP-type
peptidyl-prolyl cis-trans isomerase-like, partial (36%)
Length = 539
Score = 56.6 bits (135), Expect = 2e-08
Identities = 29/68 (42%), Positives = 43/68 (62%), Gaps = 3/68 (4%)
Frame = +2
Query: 401 GEAPFKFRLG---KGEVIEGWDIGLEGMRVGEKRRLVVPPSMTSKKDGESENIPPNSWLV 457
G P+ F +G +G V++G D+G++GMRVG +R L+VPP + G E IPPNS +
Sbjct: 149 GGTPYGFDVGQSERGTVLKGLDLGVQGMRVGGQRLLIVPPELAYGSKGVQE-IPPNSTIE 325
Query: 458 YDFELVKV 465
D EL+ +
Sbjct: 326 LDIELLSI 349
>TC206087 similar to UP|Q38931 (Q38931) Rof1, partial (50%)
Length = 1183
Score = 55.5 bits (132), Expect = 5e-08
Identities = 29/90 (32%), Positives = 50/90 (55%), Gaps = 4/90 (4%)
Frame = +3
Query: 378 GKKISINYTGKLKENGVVVESN-AGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVP 436
G + + GKL++ V V+ E PF+F++ + +VI+G D ++ M+ GE L++
Sbjct: 72 GAVVQVKLIGKLQDGTVFVKKGYVDEQPFEFKIDEEQVIDGLDQAVKNMKKGEIALLIIQ 251
Query: 437 PSMTSKKDGESE---NIPPNSWLVYDFELV 463
P G S+ N+PPNS + Y+ EL+
Sbjct: 252 PEYAFGPSGSSQELANVPPNSTVYYEVELL 341
>TC225581 homologue to UP|O04287 (O04287) Immunophilin, complete
Length = 812
Score = 55.1 bits (131), Expect = 7e-08
Identities = 32/92 (34%), Positives = 53/92 (56%), Gaps = 6/92 (6%)
Frame = +2
Query: 378 GKKISINYTGKLKENGVVVES-----NAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRR 432
G+ ++++ TG +NG + + + G+ PF F++G+G VI+GWD G+ GM++GE R
Sbjct: 134 GQNVTVHCTG-FGKNGDLSQKFWSTKDPGQDPFTFKIGQGSVIKGWDEGVLGMQIGEVAR 310
Query: 433 LVVPPSMTSKKDG-ESENIPPNSWLVYDFELV 463
L P G S I PNS L ++ E++
Sbjct: 311 LRCSPDYAYGAGGFPSWGIQPNSVLEFEIEVL 406
>TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%)
Length = 870
Score = 54.3 bits (129), Expect = 1e-07
Identities = 58/254 (22%), Positives = 97/254 (37%), Gaps = 6/254 (2%)
Frame = +3
Query: 22 NSIIETKLKTDNAFTESQIHSREAGPSDHLVDPCTVPDVGDIKKSKKKKKGKEKETKSSS 81
N + K K +++ +S S + P+ + P + S KKGK + SSS
Sbjct: 36 NEVKVQKGKEESSSDDSSSDSEDEKPAAKVAVPSKNQSAKNGTLSTLAKKGKPAASSSSS 215
Query: 82 NGDSTELDNAEQDE-PKMNMAQDLLDQEIVDNVDKEAVDDGKKDGEDRENITIETKLKTD 140
DN+++DE PK +A + + D D ++ + E K K
Sbjct: 216 ESSD---DNSDEDEAPKTKVAPAAGKNGHASTKKTQPSESSDSDSSDSDSSSDEGKSKKK 386
Query: 141 NVVADIQT-----HREAEPSDQLAVPSTVPDVGDMKKSKKKKKGKEKETKSSSNEHSTDL 195
A + T ++ E SD + S+ D D K + + ++ S+D
Sbjct: 387 PTTAKLPTLPVAPAKKVESSDDESSESSDED-NDAKPAVTAVS--KPSARAQKKVESSDS 557
Query: 196 DNAAQDEPKMNMDEDLLTGNEQNQQQPDDKKAETTDKTLPSSQVGQGQDEKPKRKRKERS 255
D+++ DE K MD D + ++P KKA K S +DE ++ K
Sbjct: 558 DDSSSDEDKGKMDIDDDDDSSDESEEPQKKKAVKNSKESSDSSEEDSEDESEEKPSKTPQ 737
Query: 256 KEKTLFAADDACIS 269
K DA +S
Sbjct: 738 KRGRDVEMVDAALS 779
>TC225580 homologue to UP|O04287 (O04287) Immunophilin, complete
Length = 631
Score = 53.5 bits (127), Expect = 2e-07
Identities = 31/92 (33%), Positives = 53/92 (56%), Gaps = 6/92 (6%)
Frame = +1
Query: 378 GKKISINYTGKLKENGVVVES-----NAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRR 432
G+ ++++ TG +NG + + + G+ PF F++G+G VI+GWD G+ GM++GE R
Sbjct: 151 GQNVTVHCTG-FGKNGDLSQKFWSTKDPGQNPFTFKIGQGSVIKGWDEGVLGMQIGEVAR 327
Query: 433 LVVPPSMTSKKDG-ESENIPPNSWLVYDFELV 463
L P G + I PNS L ++ E++
Sbjct: 328 LRCSPDYAYGAGGFPAWGIRPNSVLEFEIEVL 423
>TC216038 similar to UP|Q9IZF4 (Q9IZF4) Aphid transmission P5, partial (3%)
Length = 1754
Score = 51.6 bits (122), Expect = 7e-07
Identities = 51/212 (24%), Positives = 89/212 (41%), Gaps = 1/212 (0%)
Frame = +3
Query: 144 ADIQTHREAEPSDQLAVPSTVPDVGDMKKSKKKKKGKEKETKSSSNEHSTDLDNAAQDEP 203
AD+ + + A P +V V + SK+ K G ++++S D+ + DE
Sbjct: 666 ADVDDEDDDDYYATTAPPQSVWGVSEPHHSKEDKHGNFEDSESEE-----DILDEGDDEV 830
Query: 204 KMNMDEDLLTGNEQNQQQPDDKKAETTDKTLPSSQVGQGQDEKPKRKRKERSKEKTLFAA 263
+ D + + +P+ + + + LP + + Q K +RK+KE ++ + L A
Sbjct: 831 EEEHDHE-----PEESLKPEPEVKKDAEVPLPPPKETERQLSKKERKKKELAELEALLA- 992
Query: 264 DDACISNVVNLPQGNEHSNQDTVNGSVDNGAQDNPDGNQSEDKKVKKKKKKSKSQGSEVV 323
D N Q QD G +G + + +E K KKKKKK K+
Sbjct: 993 DFGVAPKQSNDGQDESQGAQDK-KGVETDGDGEKKENVPAESKTSKKKKKKDKASKEVKE 1169
Query: 324 NSDVPVSAE-QSSEMMKEDGNNVEDTKPSQVR 354
+ D P S++ + + NVEDT V+
Sbjct: 1170SQDQPNSSDTNNGPDLATGAENVEDTSTVDVK 1265
Score = 35.0 bits (79), Expect = 0.071
Identities = 40/150 (26%), Positives = 58/150 (38%), Gaps = 12/150 (8%)
Frame = +3
Query: 64 KKSKKKKKGKEKETKSSSNGDSTELDNAEQDEPKMNMAQDLLDQEIVDNVDKEAVDDGKK 123
KK +KKK+ E E + G + + N QDE +Q D++ V+ DG
Sbjct: 942 KKERKKKELAELEALLADFGVAPKQSNDGQDE-----SQGAQDKKGVET-------DG-- 1079
Query: 124 DGEDRENITIETKLKTDNVVADIQTHREAEPSDQLAVPST------------VPDVGDMK 171
DGE +EN+ E+K D + E DQ T V D +
Sbjct: 1080 DGEKKENVPAESKTSKKKKKKDKASKEVKESQDQPNSSDTNNGPDLATGAENVEDTSTVD 1259
Query: 172 KSKKKKKGKEKETKSSSNEHSTDLDNAAQD 201
++ KK + K SS E AAQ+
Sbjct: 1260 VKERLKKVASTKKKKSSKEMDAAARAAAQE 1349
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.302 0.124 0.332
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,656,655
Number of Sequences: 63676
Number of extensions: 164749
Number of successful extensions: 5110
Number of sequences better than 10.0: 156
Number of HSP's better than 10.0 without gapping: 1571
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3581
length of query: 466
length of database: 12,639,632
effective HSP length: 101
effective length of query: 365
effective length of database: 6,208,356
effective search space: 2266049940
effective search space used: 2266049940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146527.7


