

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146527.1 - phase: 0
(468 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
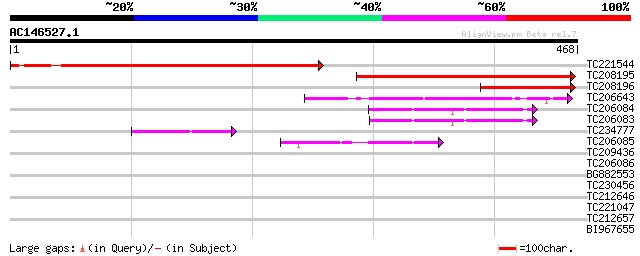
Sequences producing significant alignments: (bits) Value
TC221544 403 e-113
TC208195 311 3e-85
TC208196 135 4e-32
TC206643 similar to GB|AAO44043.1|28466869|BT004777 At1g06060 {A... 67 2e-11
TC206084 similar to UP|Q84WK5 (Q84WK5) At1g61150, partial (92%) 61 1e-09
TC206083 similar to UP|Q84WK5 (Q84WK5) At1g61150, partial (72%) 58 8e-09
TC234777 similar to GB|AAC13563.1|3046995|AF056717 ash2l2 {Homo ... 47 2e-05
TC206085 similar to UP|Q84WK5 (Q84WK5) At1g61150, partial (51%) 44 1e-04
TC209436 32 0.79
TC206086 similar to UP|Q84WK5 (Q84WK5) At1g61150, partial (33%) 31 1.4
BG882553 similar to GP|1405348|dbj zinc-finger DNA-binding prote... 30 3.0
TC230456 similar to UP|Q94A50 (Q94A50) AT3g55070/T15C9_70, parti... 30 3.0
TC212646 29 3.9
TC221047 similar to UP|Q6K784 (Q6K784) IgE autoantigen-like, par... 29 3.9
TC212657 similar to UP|Q8GTM3 (Q8GTM3) Chlorophyllase 2 , parti... 29 5.1
BI967655 weakly similar to GP|169917|gb|AA auxin-regulated prote... 28 6.7
>TC221544
Length = 777
Score = 403 bits (1036), Expect = e-113
Identities = 200/261 (76%), Positives = 226/261 (85%), Gaps = 2/261 (0%)
Frame = +2
Query: 1 MKETNGKNTIEKPDQDQNQDLGFHFLELSR-RKAKLDSEDFEVDDEEKEESPSELNTINS 59
M E+NG + + ++QDLG +FLE R R+ K ++EE+EE+P ELNTINS
Sbjct: 26 MSESNGNS---RRRNVEDQDLGLYFLEACRGREMK--------EEEEEEEAPRELNTINS 172
Query: 60 SGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIG 119
SGGFVVV+TDKLSVKYTSVNLHGHDVGVIQANK AP KR++YYFEI VKDAGVKGQ++IG
Sbjct: 173 SGGFVVVSTDKLSVKYTSVNLHGHDVGVIQANKPAPTKRLMYYFEIHVKDAGVKGQIAIG 352
Query: 120 FTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGK-GEAFGPTYTTGDIVGAGINYAAQEF 178
FT+E+FKMRRQPGWEANSCGYHGDDG LYRG GK GE FGPTYT+GD+VGAGINYAAQEF
Sbjct: 353 FTSETFKMRRQPGWEANSCGYHGDDGLLYRGHGKGGEPFGPTYTSGDVVGAGINYAAQEF 532
Query: 179 FFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIK 238
FFTKNG VVG+V+K+MKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKE+E QERMKQQ+K
Sbjct: 533 FFTKNGHVVGSVYKDMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEFETQERMKQQLK 712
Query: 239 IEEVPVSPNASYGIVRSYLLH 259
IEE+ V PN SYGIVRSYLLH
Sbjct: 713 IEEISVPPNVSYGIVRSYLLH 775
>TC208195
Length = 797
Score = 311 bits (798), Expect = 3e-85
Identities = 155/181 (85%), Positives = 169/181 (92%)
Frame = +2
Query: 287 GIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIEL 346
GID+QE TYALNHRKTLRQLIR+G+ID AFGKLREWYPQI EDNTSA CFLLHCQKFIEL
Sbjct: 2 GIDEQEITYALNHRKTLRQLIRNGDIDVAFGKLREWYPQIVEDNTSATCFLLHCQKFIEL 181
Query: 347 VRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVA 406
VRVGALEEAVKYGR+ELSSF+ L +F+D+VQDCVALLAYERPLES+VGYLLKDSQREVVA
Sbjct: 182 VRVGALEEAVKYGRMELSSFYDLPVFKDLVQDCVALLAYERPLESSVGYLLKDSQREVVA 361
Query: 407 DAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACCLERRSLSGEQGEAFQLQRVLSSGK 466
D VNAMILSTN N+K +K+CLHS LERLLRQLTACCL RRSL+GEQGEAFQLQRVLSS +
Sbjct: 362 DTVNAMILSTNLNMKDSKHCLHSYLERLLRQLTACCLVRRSLNGEQGEAFQLQRVLSSSR 541
Query: 467 R 467
R
Sbjct: 542 R 544
>TC208196
Length = 566
Score = 135 bits (340), Expect = 4e-32
Identities = 68/79 (86%), Positives = 74/79 (93%)
Frame = +1
Query: 389 LESAVGYLLKDSQREVVADAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACCLERRSL 448
LES+VGYLLKDSQREVVAD VNAMILS NPN+K +K+CLHS LERLLRQLTACCLERRSL
Sbjct: 1 LESSVGYLLKDSQREVVADTVNAMILSANPNMKDSKHCLHSYLERLLRQLTACCLERRSL 180
Query: 449 SGEQGEAFQLQRVLSSGKR 467
+GEQGEAFQLQRVLSS +R
Sbjct: 181 NGEQGEAFQLQRVLSSSRR 237
>TC206643 similar to GB|AAO44043.1|28466869|BT004777 At1g06060 {Arabidopsis
thaliana;} , partial (76%)
Length = 1277
Score = 67.0 bits (162), Expect = 2e-11
Identities = 62/223 (27%), Positives = 110/223 (48%), Gaps = 2/223 (0%)
Frame = +1
Query: 244 VSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYALNHRKTL 303
V+ N + IV SYL+H Y++++ SF + +T P + ++D ++ RK +
Sbjct: 136 VNDNDIHNIVLSYLIHNCYKESVESFIACTGATQPA------DYLED------MDKRKRI 279
Query: 304 RQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVKYGRIEL 363
+G A + I E+N LL F+ELV EA+++ + +L
Sbjct: 280 FHFALEGNALKAIELTEQLAKDILENNKDLQFDLLSLH-FVELVCSRKCTEALEFAQTKL 456
Query: 364 SSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILSTNPNIKVT 423
F + + ++D +ALLAY+ P +S + +LL R+ VAD++N IL+ + N+
Sbjct: 457 GPFGKEPKYMEKLEDFMALLAYKEPEKSPMFHLLSLEYRQQVADSLNRAILA-HLNLP-- 627
Query: 424 KNCLHSNLERLLRQLTAC--CLERRSLSGEQGEAFQLQRVLSS 464
++ +ERL++Q T CL + +G+ G F L+ L S
Sbjct: 628 ---SYTAMERLIQQATVVRQCLSQE--AGKDGPPFSLKDFLKS 741
>TC206084 similar to UP|Q84WK5 (Q84WK5) At1g61150, partial (92%)
Length = 1064
Score = 60.8 bits (146), Expect = 1e-09
Identities = 48/141 (34%), Positives = 77/141 (54%), Gaps = 2/141 (1%)
Frame = +3
Query: 297 LNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAV 356
+ R +++ ++ G ++ A K+ + P+I + N + F L Q+ IEL+R G +EEA+
Sbjct: 477 ITDRMAVKKAVQSGNVEDAIEKVNDLNPEILDTNPQ-LFFHLQQQRLIELIRNGKVEEAL 653
Query: 357 KYGRIELS--SFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMIL 414
++ + EL+ S E++ + VALLA+E VG LL SQR A VNA IL
Sbjct: 654 EFAQEELAPRGEENQSFLEEL-ERTVALLAFEDVSNCPVGELLDISQRLKTASEVNAAIL 830
Query: 415 STNPNIKVTKNCLHSNLERLL 435
++ + K K L S L+ LL
Sbjct: 831 TSQSHEKDPK--LPSLLKMLL 887
>TC206083 similar to UP|Q84WK5 (Q84WK5) At1g61150, partial (72%)
Length = 789
Score = 58.2 bits (139), Expect = 8e-09
Identities = 48/140 (34%), Positives = 74/140 (52%), Gaps = 2/140 (1%)
Frame = +2
Query: 298 NHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVK 357
N + ++ G ++ A K+ + P+I + N + F L Q+ IEL+R G +EEA++
Sbjct: 86 NRSHGCKXXVQSGNVEDAIEKVNDLNPEILDTNPQ-LFFHLQQQRLIELIRNGKVEEALE 262
Query: 358 YGRIELS--SFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILS 415
+ + EL+ S E++ + VALLA+E VG LL SQR A VNA IL+
Sbjct: 263 FAQEELAPRGEENQSFLEEL-ERTVALLAFEDVSNCPVGELLDISQRLKTASEVNAAILT 439
Query: 416 TNPNIKVTKNCLHSNLERLL 435
+ + K K L S L+ LL
Sbjct: 440 SQSHEKDPK--LPSLLKMLL 493
>TC234777 similar to GB|AAC13563.1|3046995|AF056717 ash2l2 {Homo sapiens;} ,
partial (8%)
Length = 893
Score = 47.0 bits (110), Expect = 2e-05
Identities = 29/88 (32%), Positives = 44/88 (49%), Gaps = 1/88 (1%)
Frame = +2
Query: 101 YYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPT 160
+YFEI V G G +G++TE ++ G++ NS GY DG + E +G
Sbjct: 233 WYFEIRVVKLGESGHTRLGWSTEKGDLQAPVGYDGNSFGYRDIDGSKIH-EALREKYGEE 409
Query: 161 -YTTGDIVGAGINYAAQEFFFTKNGQVV 187
Y GD++G IN E + K+ Q+V
Sbjct: 410 GYKEGDVIGFYINLPDGEQYAPKSSQLV 493
>TC206085 similar to UP|Q84WK5 (Q84WK5) At1g61150, partial (51%)
Length = 753
Score = 44.3 bits (103), Expect = 1e-04
Identities = 35/142 (24%), Positives = 65/142 (45%), Gaps = 7/142 (4%)
Frame = +3
Query: 224 LKEYEAQERMKQQI-------KIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRST 276
L E EA K+ I K+ +V + +V ++L+ GY + F S T
Sbjct: 369 LAEIEAMAASKKLITREEWEKKLSDVKIRKEDMNKLVMNFLVTEGYVEAAEKFRMES-GT 545
Query: 277 IPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCF 336
P I++ + R +++ ++ G ++ A K+ + P+I + N + F
Sbjct: 546 EPDIDLA------------TITDRMAVKKAVQSGNVEDAIEKVNDLNPEILDTNPQ-LFF 686
Query: 337 LLHCQKFIELVRVGALEEAVKY 358
L Q+ IEL+R G +EEA+++
Sbjct: 687 HLQQQRLIELIRNGKVEEALEF 752
>TC209436
Length = 892
Score = 31.6 bits (70), Expect = 0.79
Identities = 17/44 (38%), Positives = 23/44 (51%), Gaps = 1/44 (2%)
Frame = -2
Query: 104 EIFVKDAG-VKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGF 146
+ +K AG V +G + SFK QP +SC + GDDGF
Sbjct: 492 DCILKAAGSVMASTDLGSASASFKSVLQPHSRISSCRHFGDDGF 361
>TC206086 similar to UP|Q84WK5 (Q84WK5) At1g61150, partial (33%)
Length = 560
Score = 30.8 bits (68), Expect = 1.4
Identities = 16/41 (39%), Positives = 27/41 (65%)
Frame = +3
Query: 324 PQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVKYGRIELS 364
P+I + N + F L Q+ IEL+R G +EEA+++ + EL+
Sbjct: 435 PEILDTNPQ-LFFHLQQQRLIELIRNGKVEEALEFAQEELA 554
>BG882553 similar to GP|1405348|dbj zinc-finger DNA-binding protein {Homo
sapiens}, partial (1%)
Length = 399
Score = 29.6 bits (65), Expect = 3.0
Identities = 17/53 (32%), Positives = 26/53 (48%), Gaps = 1/53 (1%)
Frame = +2
Query: 176 QEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVN-FGQKPFTFDLKEY 227
+E FF+ +G G + K+ K P+ V S + HV+ FG + FT Y
Sbjct: 107 EEQFFSPSGSSGGNINKQQKSSPSPSGVVASSSRVFHVDKFGSRSFTSRTPSY 265
>TC230456 similar to UP|Q94A50 (Q94A50) AT3g55070/T15C9_70, partial (72%)
Length = 907
Score = 29.6 bits (65), Expect = 3.0
Identities = 35/152 (23%), Positives = 66/152 (43%), Gaps = 1/152 (0%)
Frame = +3
Query: 252 IVRSYLLHYGYEDTLNSFDEASR-STIPPINIVQENGIDDQETTYALNHRKTLRQLIRDG 310
I+ Y+L Y DT E+S + I++ QE K + +++
Sbjct: 351 ILVDYMLRMSYYDTAVKLAESSNLQDLVDIDVFQE--------------AKKVIDALQNK 488
Query: 311 EIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLS 370
++ A + ++ + S M F L Q+FIELVR A+ Y L+ +
Sbjct: 489 DVAPALAWCADNKSRLKKSK-SKMEFQLRLQEFIELVRAENNLRAITYATKYLAPWGATH 665
Query: 371 LFEDIVQDCVALLAYERPLESAVGYLLKDSQR 402
+ E +Q +A LA++R E + +L ++++
Sbjct: 666 MKE--LQRVIATLAFKRDTECSTYKVLFEAKQ 755
>TC212646
Length = 434
Score = 29.3 bits (64), Expect = 3.9
Identities = 13/29 (44%), Positives = 17/29 (57%)
Frame = -1
Query: 118 IGFTTESFKMRRQPGWEANSCGYHGDDGF 146
+G + SFK QP +SC + GDDGF
Sbjct: 242 LGSASASFKSVLQPHSRISSCRHFGDDGF 156
>TC221047 similar to UP|Q6K784 (Q6K784) IgE autoantigen-like, partial (61%)
Length = 763
Score = 29.3 bits (64), Expect = 3.9
Identities = 23/77 (29%), Positives = 35/77 (44%)
Frame = +2
Query: 28 LSRRKAKLDSEDFEVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGV 87
+ ++K+KL VDDEEKE + I + F + T K + + S HG G
Sbjct: 74 MDKKKSKLVGI---VDDEEKEAQKTREIRIERTDEFGRILTPKEAFRMISHKFHGKGPG- 241
Query: 88 IQANKVAPMKRIVYYFE 104
K+ KR+ Y+E
Sbjct: 242 ----KMKQEKRMKQYYE 280
>TC212657 similar to UP|Q8GTM3 (Q8GTM3) Chlorophyllase 2 , partial (52%)
Length = 811
Score = 28.9 bits (63), Expect = 5.1
Identities = 21/70 (30%), Positives = 30/70 (42%), Gaps = 2/70 (2%)
Frame = +3
Query: 171 INYAAQEFFFTKNGQVVGTVFKEMK-GPLFPTVAVHSQNEEVHVNFGQKP-FTFDLKEYE 228
+ Y F F V+G+ E+K PLFP A N E N +KP + F K+Y
Sbjct: 183 LTYVPNSFDFDMAVMVIGSGLGEVKRNPLFPPCAPKGVNHENFFNECKKPAWYFVAKDYG 362
Query: 229 AQERMKQQIK 238
+ + K
Sbjct: 363 HSDMLDDDTK 392
>BI967655 weakly similar to GP|169917|gb|AA auxin-regulated protein {Glycine
max}, partial (53%)
Length = 250
Score = 28.5 bits (62), Expect = 6.7
Identities = 10/23 (43%), Positives = 16/23 (69%)
Frame = +2
Query: 405 VADAVNAMILSTNPNIKVTKNCL 427
+ DA+N +++ TNPN +NCL
Sbjct: 23 IHDAMNTLLVHTNPNSSPPRNCL 91
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,278,901
Number of Sequences: 63676
Number of extensions: 204684
Number of successful extensions: 1065
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 1056
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1061
length of query: 468
length of database: 12,639,632
effective HSP length: 101
effective length of query: 367
effective length of database: 6,208,356
effective search space: 2278466652
effective search space used: 2278466652
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146527.1


