

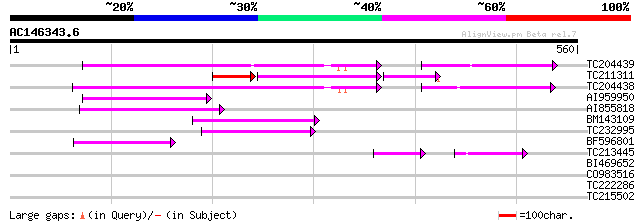
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.6 + phase: 0 /pseudo
(560 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 90 2e-18
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 44 9e-15
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 76 3e-14
AI959950 71 1e-12
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 61 2e-09
BM143109 57 2e-08
TC232995 56 4e-08
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 50 3e-06
TC213445 39 7e-04
BI469652 weakly similar to GP|18149115|dbj| reverse transcriptas... 36 0.052
CO983516 35 0.089
TC222286 similar to UP|ML10_ARATH (Q9FKY5) MLO-like protein 10 (... 28 8.3
TC215502 weakly similar to UP|Q8LL74 (Q8LL74) Cytochrome P450 mo... 28 8.3
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 90.1 bits (222), Expect = 2e-18
Identities = 102/302 (33%), Positives = 147/302 (47%), Gaps = 7/302 (2%)
Frame = +2
Query: 73 RNFLSLKETKFGT*FQIIKAKPSLEQDGFSKTSLMKKVRLSETKQGWLLKATTNRKVLTM 132
+N+ + K K G+*F ++ L G S+T MKKV ET+ WLLKAT KV T+
Sbjct: 3257 KNWSNSKGMKSGS*FLGLRELM*LAPSGSSRTKPMKKVS*PETRPDWLLKATLRLKV*TL 3436
Query: 133 MRLLHQLQG*RPSEFFLHILHIKALNFFKWM*KVHF*MVF*MRKFM*ANLLTS*MKKNQI 192
MRLL QL PS+++L L + +WM*+ HF*M *M+K M ++ + QI
Sbjct: 3437 MRLLPQLLDLSPSDYYLV*LVSSNSSCTRWM*RAHF*MDT*MKKSMWSSQRDLQTRLIQI 3616
Query: 193 MFLNSLKHFMVLNKLPELGMID-LSHF*LKMDFQEEK*IPHFSEKLITLTYS*FKFMWMI 251
M+ S + M +KL ELGM S K +EE P S K++ T ++M M
Sbjct: 3617 MYTGSRRLSMD*SKLQELGMKG*QSSLLSKGIGREELTRPSLSNKMLK-T**LHRYMLMT 3793
Query: 252 SFLVQPKRKWMKIFPISCKMNLR*A**VNLVFSLVYKSNNIQMEFS*VKKNMSMTFLKSI 311
L + + I C +NLR* + + +K + + +S K M T +S+
Sbjct: 3794 LCLEGCRMRCFDILFNRCNLNLR*VLLES*LIFWDFK*SRWRTPYSSHKAGMQRTLSRSL 3973
Query: 312 K*MR*KSW-SPIC---IHLQV*I--RMRVENLFLKKNI*A**AHYYI*LLADLA*CLQ*V 365
W P+ +HL * RM+ + +K A** YYI* LAD +Q*V
Sbjct: 3974 ------GWRMPVIKGHLHLLT*SCQRMKQAPVLIKVCTEA**GAYYI*QLADPTSPMQ*V 4135
Query: 366 YV 367
+V
Sbjct: 4136 FV 4141
Score = 45.1 bits (105), Expect = 9e-05
Identities = 40/135 (29%), Positives = 70/135 (51%)
Frame = +3
Query: 407 LL*C*LCWRQS*EKEYKWLMSIPGTSIDWMVLQKAKHHCTLNNLS*SVCICSKLLLSSFM 466
+L*C*L W+ *+K++ W M + G +MV Q+A+ + + S C K L ++ +
Sbjct: 4266 VL*C*LGWKCR*QKKHFWWMLLFGKQPYFMVQQEAELCVPIYSRSRVYC-SRKQLFTASL 4442
Query: 467 DQKSVGRLLAKVHKCSYFV**YKCN*FFKKSYSTF*V*AY*YKSSFH*RPCSEKGYYIKL 526
D+ + + +* ++C *+F KS ST A+*+ +S + R C + +
Sbjct: 4443 DEADAEGVQCRTRCHDIVL*QHECY*YF*KSCSTQQNQAH*H*TSLYQRSC***SDHTEA 4622
Query: 527 C*YRKSTS*HFHKTF 541
C*+ + S +FHK F
Sbjct: 4623 C*H*GTNSRYFHKGF 4667
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 43.9 bits (102), Expect(3) = 9e-15
Identities = 41/123 (33%), Positives = 60/123 (48%)
Frame = +1
Query: 245 FKFMWMISFLVQPKRKWMKIFPISCKMNLR*A**VNLVFSLVYKSNNIQMEFS*VKKNMS 304
F M M LVQP++ + F +M+L+ * V+ S +KS M F +K+N+
Sbjct: 496 FISMLMT*SLVQPQKGCARSFLS**RMDLKRV*KVS*SSS*DFKSFRKFMGFLSIKRNIQ 675
Query: 305 MTFLKSIK*MR*KSWSPICIHLQV*IRMRVENLFLKKNI*A**AHYYI*LLADLA*CLQ* 364
K + M+ W P+CI Q RMR + K++I * Y+I*LL D CL
Sbjct: 676 SPI*KGSEWMKPNLWQPLCIVPQSLTRMRKVIILHKRSIVV*LILYHI*LLVDQILCLSF 855
Query: 365 VYV 367
+V
Sbjct: 856 AFV 864
Score = 39.7 bits (91), Expect(3) = 9e-15
Identities = 23/42 (54%), Positives = 27/42 (63%)
Frame = +3
Query: 201 FMVLNKLPELGMIDLSHF*LKMDFQEEK*IPHFSEKLITLTY 242
FMV NKL ELGM HF* +MD EE PH+SE+L T+
Sbjct: 363 FMV*NKL*ELGMKG*VHF*FQMDSPEE*RTPHYSERLKKETF 488
Score = 34.3 bits (77), Expect(3) = 9e-15
Identities = 25/58 (43%), Positives = 34/58 (58%), Gaps = 2/58 (3%)
Frame = +2
Query: 370 MCKRISFNCN*KDF*ISSRYY*S*PLV**RF*F*SCSLL*C*LCWRQS*EKEY--KWL 425
+ K S C+*KD IS Y*S +V* + * *S +L*C CW S ++E+ KW+
Sbjct: 878 LSKNFSCYCS*KDLKISCWNY*SLSMV*EKV*V*SFRIL*CLFCW**SRKEEH**KWM 1051
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 76.3 bits (186), Expect = 3e-14
Identities = 97/311 (31%), Positives = 142/311 (45%), Gaps = 6/311 (1%)
Frame = +2
Query: 63 LMTLGLRP*GRNFLSLKETKFGT*FQIIKAKPSLEQDGFSKTSLMKKVRLSETKQGWLLK 122
LM+ G +N+ + K KFG+*F + L G S+T MKKV ET+ LLK
Sbjct: 3230 LMSSGSMLCKKNWSNSKGMKFGS*FLDPRELM*LAPSGSSRTKPMKKVL*PETRPDLLLK 3409
Query: 123 ATTNRKVLTMMRLLHQLQG*RPSEFFLHILHIKALNFFKWM*KVHF*MVF*MRKFM*ANL 182
AT KV T+M+L L PS+ +L L + +WM*+ F*M *M+K M ++
Sbjct: 3410 ATLRLKV*TLMKLSPLLLDLSPSDCYLV*LASSNSSCTRWM*RARF*MDT*MKKPMWSSQ 3589
Query: 183 LTS*MKKNQIMFLNSLKHFMVLNKLPELGMIDLSHF*LKMDFQEEK*IPHFSEKLITLTY 242
*++ QIM+ S + M +KL ELGM L E+ + T
Sbjct: 3590 RDL*IQLIQIMYTGSRRLSMD*SKLQELGMKG*QSSLLSKGIGREELTRLSLSNKMLKT* 3769
Query: 243 S*FKFMWMISFLVQPKRKWMKIFPISCKMNLR*A**VNLVFSLVYKSNNIQMEFS*VKKN 302
* ++M M L + + I C +NLR* + + K + + +S K +
Sbjct: 3770 **HRYMLMTLCLEGCRMRCFDILSNRCNLNLR*VLLES*LIFWDSK*SRWKTPYSSHKAS 3949
Query: 303 MSMTFLKSIK*MR*KSWSPICI----HLQV*I--RMRVENLFLKKNI*A**AHYYI*LLA 356
M T +S+ W I HL * +M++ + +K A* YYI* LA
Sbjct: 3950 MQRTLSRSL------GWKMPAIKEHLHLLT*SCQKMKLAPVLIKVCTEA*LGAYYI*QLA 4111
Query: 357 DLA*CLQ*VYV 367
DL +Q*V+V
Sbjct: 4112 DLTSPMQ*VFV 4144
Score = 47.8 bits (112), Expect = 1e-05
Identities = 43/134 (32%), Positives = 71/134 (52%), Gaps = 1/134 (0%)
Frame = +3
Query: 407 LL*C*LCWRQS*EKEYKWLMSIPGTSIDWMVLQKAKHHCTLNNLS*SVCICS-KLLLSSF 465
+L*C*L W+ *+K++ W M + G +MV Q+A+ + *S CS K L ++
Sbjct: 4269 VL*C*LGWKCR*QKKHFWWMFLFGNQSYFMVQQEAE--LCVPIYC*SRVYCSRKQLFTTS 4442
Query: 466 MDQKSVGRLLAKVHKCSYFV**YKCN*FFKKSYSTF*V*AY*YKSSFH*RPCSEKGYYIK 525
+D+ + + +* ++C *+F KS ST A+*+ +S +*R C Y+
Sbjct: 4443 LDEADAEGVQCRTRCHDIVL*QHECY*YF*KSCSTQQNQAH*H*TSLY*RSC***SYHTG 4622
Query: 526 LC*YRKSTS*HFHK 539
C*+ + S +FHK
Sbjct: 4623 AC*H*GTNSRYFHK 4664
>AI959950
Length = 466
Score = 70.9 bits (172), Expect = 1e-12
Identities = 53/127 (41%), Positives = 67/127 (52%)
Frame = -2
Query: 73 RNFLSLKETKFGT*FQIIKAKPSLEQDGFSKTSLMKKVRLSETKQGWLLKATTNRKVLTM 132
+N +S K + K + LE +G+ T+ + VRL +TKQ LLK T NRKV T
Sbjct: 381 KNLISFKRIMSRSSLNYQKERR*LE*NGYFVTN*TRMVRL*DTKQD*LLKVTHNRKV*TT 202
Query: 133 MRLLHQLQG*RPSEFFLHILHIKALNFFKWM*KVHF*MVF*MRKFM*ANLLTS*MKKNQI 192
+ LH L *+ + H+ I + KWM*KVHF*M RKFM N L MK
Sbjct: 201 QKPLHLLHV*K*YASYFHLQPIVI*SCIKWM*KVHF*MA*SKRKFMLNNRLDLKMKPFIN 22
Query: 193 MFLNSLK 199
MFLNS K
Sbjct: 21 MFLNSTK 1
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 60.8 bits (146), Expect = 2e-09
Identities = 52/143 (36%), Positives = 73/143 (50%)
Frame = -1
Query: 70 P*GRNFLSLKETKFGT*FQIIKAKPSLEQDGFSKTSLMKKVRLSETKQGWLLKATTNRKV 129
P +N ++LKE G * + +K S EQ+GF + + M L E + K ++
Sbjct: 457 PCKKN*INLKEIMCGN**KNLKIILS*EQNGFLEIN*MNMA*LLEIRLD**QKGIIKKRE 278
Query: 130 LTMMRLLHQLQG*RPSEFFLHILHIKALNFFKWM*KVHF*MVF*MRKFM*ANLLTS*MKK 189
TM + + QLQ *+ E F H+ LN KWM KV F*MV +K+M N +
Sbjct: 277 *TMKKHMLQLQD*KSLECF*HMYP**ILNSIKWMLKVLF*MV*FKKKYMLNNPQALKSRI 98
Query: 190 NQIMFLNSLKHFMVLNKLPELGM 212
NQ+MF+N + FMV NK GM
Sbjct: 97 NQLMFINCKRLFMV*NKPLGRGM 29
>BM143109
Length = 415
Score = 57.0 bits (136), Expect = 2e-08
Identities = 49/126 (38%), Positives = 69/126 (53%)
Frame = +2
Query: 181 NLLTS*MKKNQIMFLNSLKHFMVLNKLPELGMIDLSHF*LKMDFQEEK*IPHFSEKLITL 240
NLL +K+ IM LN + +M NK LGM +F FQ+ + I F K +
Sbjct: 5 NLL*GKTQKSLIMSLN*KRFYMD*NKPLGLGMNF*VNFF*TRVFQKVRLILTFLFKRN*M 184
Query: 241 TYS*FKFMWMISFLVQPKRKWMKIFPISCKMNLR*A**VNLVFSLVYKSNNIQMEFS*VK 300
YS*+++M MI FLVQ + K F CKMNL+ * V+ F L YKS+ +ME+ V
Sbjct: 185 IYS*YRYMLMILFLVQLMILFAKNFLKICKMNLKCQ*CVS*TFFLDYKSSKQRMEYLSVN 364
Query: 301 KNMSMT 306
+N++ T
Sbjct: 365 QNIAKT 382
>TC232995
Length = 1009
Score = 56.2 bits (134), Expect = 4e-08
Identities = 42/113 (37%), Positives = 58/113 (51%)
Frame = +3
Query: 190 NQIMFLNSLKHFMVLNKLPELGMIDLSHF*LKMDFQEEK*IPHFSEKLITLTYS*FKFMW 249
NQ MF+N + FMV NK GM D F LK + E K IPH+S + + + FK+M
Sbjct: 36 NQTMFINYKRLFMV*NKPLGHGMND*VIFFLKKNSPEVKWIPHYS*RESIMIFCWFKYML 215
Query: 250 MISFLVQPKRKWMKIFPISCKMNLR*A**VNLVFSLVYKSNNIQMEFS*VKKN 302
MI FL + FP+ CK+NL+ * N YKS+ + +S + N
Sbjct: 216 MI*FLDPLMIHCARSFPLICKVNLKCQ*WEN*STFWDYKSSKLNKVYSSINPN 374
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 49.7 bits (117), Expect = 3e-06
Identities = 37/100 (37%), Positives = 52/100 (52%)
Frame = +1
Query: 64 MTLGLRP*GRNFLSLKETKFGT*FQIIKAKPSLEQDGFSKTSLMKKVRLSETKQGWLLKA 123
M +G +N +LKET +G * + +K LEQ+GF + + M V L E K G K
Sbjct: 34 MIIGSLSCKKN*TNLKETMYGN**KNLKIILLLEQNGFLEIN*MNMV*LLEIKPG**RKD 213
Query: 124 TTNRKVLTMMRLLHQLQG*RPSEFFLHILHIKALNFFKWM 163
++ TM + + LQ *+P E F H+ LNF KWM
Sbjct: 214 IIKKRE*TMKKHMLLLQD*KPLECFWHMHP**TLNFIKWM 333
>TC213445
Length = 705
Score = 39.3 bits (90), Expect(2) = 7e-04
Identities = 29/72 (40%), Positives = 41/72 (56%)
Frame = +3
Query: 440 KAKHHCTLNNLS*SVCICSKLLLSSFMDQKSVGRLLAKVHKCSYFV**YKCN*FFKKSYS 499
KAK C +N S + C KLL ++ +D+ + L + +Y +* YKCN* +KSYS
Sbjct: 471 KAK*CCLINCRS-RIYFC*KLLCTNLLDETTTF*LWFET*SYTYPM*QYKCN*SIQKSYS 647
Query: 500 TF*V*AY*YKSS 511
* AY* K+S
Sbjct: 648 VL*NKAY*NKAS 683
Score = 21.9 bits (45), Expect(2) = 7e-04
Identities = 19/51 (37%), Positives = 27/51 (52%)
Frame = +1
Query: 360 *CLQ*VYVLNMCKRISFNCN*KDF*ISSRYY*S*PLV**RF*F*SCSLL*C 410
*CL + + +RIS C+*K+ I +Y S +V * F * +L*C
Sbjct: 247 *CLYVCKISSKSQRISPKCH*KNNEIFVGHYKSRVMVS*EFLL*LSRIL*C 399
>BI469652 weakly similar to GP|18149115|dbj| reverse transcriptase {Silene
noctiflora}, partial (60%)
Length = 427
Score = 35.8 bits (81), Expect = 0.052
Identities = 28/61 (45%), Positives = 32/61 (51%)
Frame = +1
Query: 154 IKALNFFKWM*KVHF*MVF*MRKFM*ANLLTS*MKKNQIMFLNSLKHFMVLNKLPELGMI 213
IK FFKW+ KV F*M RK M NL T * +F N + MV N+ LGMI
Sbjct: 235 IKT*IFFKWILKVVF*MTLLKRKCMSDNLQTL*TIHF*TIFSNFKRLHMV*NRHLMLGMI 414
Query: 214 D 214
D
Sbjct: 415 D 417
>CO983516
Length = 724
Score = 35.0 bits (79), Expect = 0.089
Identities = 26/69 (37%), Positives = 39/69 (55%)
Frame = +3
Query: 144 PSEFFLHILHIKALNFFKWM*KVHF*MVF*MRKFM*ANLLTS*MKKNQIMFLNSLKHFMV 203
PS+ +L L + +WM*+ F*M *M+K M ++ *++ QIM+ S + M
Sbjct: 390 PSDCYLV*LASSNSSCTRWM*RARF*MDT*MKKSMWSSQRDL*IQLIQIMYTGSRRLSMD 569
Query: 204 LNKLPELGM 212
+KL ELGM
Sbjct: 570 *SKLQELGM 596
>TC222286 similar to UP|ML10_ARATH (Q9FKY5) MLO-like protein 10 (AtMlo10),
partial (25%)
Length = 731
Score = 28.5 bits (62), Expect = 8.3
Identities = 11/27 (40%), Positives = 17/27 (62%)
Frame = +1
Query: 317 KSWSPICIHLQV*IRMRVENLFLKKNI 343
KSW IC H+++ IR + +K+NI
Sbjct: 406 KSWQTICYHVRINIRRTKRHQIVKRNI 486
>TC215502 weakly similar to UP|Q8LL74 (Q8LL74) Cytochrome P450 monooxygenase
CYP72A16, partial (27%)
Length = 991
Score = 28.5 bits (62), Expect = 8.3
Identities = 16/40 (40%), Positives = 24/40 (60%), Gaps = 3/40 (7%)
Frame = +3
Query: 103 KTSLMKKVRLSETKQGWLLKATTN---RKVLTMMRLLHQL 139
+T LM K++ + WLL TTN +K+ T +R LH+L
Sbjct: 750 QTGLMMKLQNAYIPGWWLLPTTTNKRMKKIDTEIRALHKL 869
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.379 0.169 0.641
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,812,223
Number of Sequences: 63676
Number of extensions: 332222
Number of successful extensions: 4820
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1484
Number of HSP's successfully gapped in prelim test: 155
Number of HSP's that attempted gapping in prelim test: 3296
Number of HSP's gapped (non-prelim): 1717
length of query: 560
length of database: 12,639,632
effective HSP length: 102
effective length of query: 458
effective length of database: 6,144,680
effective search space: 2814263440
effective search space used: 2814263440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 13 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 35 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146343.6


