

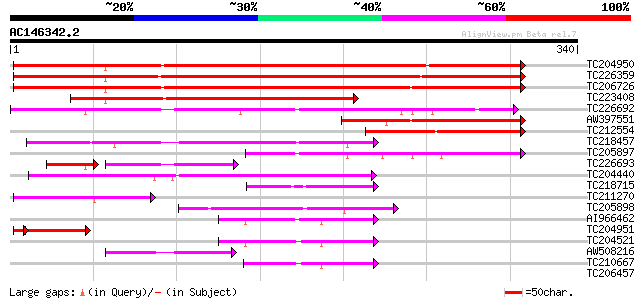
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146342.2 + phase: 0
(340 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204950 similar to UP|Q945P3 (Q945P3) At1g23740/F5O8_27, partia... 289 1e-78
TC226359 similar to UP|O23939 (O23939) Ripening-induced protein ... 285 3e-77
TC206726 similar to UP|O23939 (O23939) Ripening-induced protein ... 274 5e-74
TC223408 similar to UP|Q43677 (Q43677) Auxin-induced protein, pa... 162 3e-40
TC226692 similar to UP|Q9AYU1 (Q9AYU1) Quinone-oxidoreductase QR... 138 3e-33
AW397551 similar to PIR|T10824|T10 auxin-induced protein (clone ... 99 2e-21
TC212554 similar to UP|Q43677 (Q43677) Auxin-induced protein, pa... 87 8e-18
TC218457 similar to PIR|T05166|T05166 quinone reductase homolog ... 82 3e-16
TC205897 similar to UP|Q9SXZ7 (Q9SXZ7) NADPH oxidoreductase homo... 71 8e-13
TC226693 weakly similar to UP|Q6T7C8 (Q6T7C8) Fiber quinone-oxid... 54 9e-10
TC204440 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehy... 60 1e-09
TC218715 similar to UP|Q84N44 (Q84N44) Oxidoreductase, partial (... 48 7e-06
TC211270 similar to UP|Q84N44 (Q84N44) Oxidoreductase, partial (... 46 3e-05
TC205898 similar to UP|Q9SXZ7 (Q9SXZ7) NADPH oxidoreductase homo... 44 1e-04
AI966462 44 1e-04
TC204951 similar to UP|Q8LEB8 (Q8LEB8) Quinone oxidoreductase-li... 41 3e-04
TC204521 similar to UP|Q9SLN8 (Q9SLN8) Allyl alcohol dehydrogena... 42 3e-04
AW508216 similar to PIR|T49047|T49 quinone reductase-like protei... 41 7e-04
TC210667 similar to UP|Q6WAU0 (Q6WAU0) (+)-pulegone reductase, p... 41 9e-04
TC206457 weakly similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannit... 40 0.001
>TC204950 similar to UP|Q945P3 (Q945P3) At1g23740/F5O8_27, partial (80%)
Length = 1560
Score = 289 bits (740), Expect = 1e-78
Identities = 146/310 (47%), Positives = 210/310 (67%), Gaps = 3/310 (0%)
Frame = +1
Query: 3 KAWFYEEYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FPA 59
KAW Y EYG +VLKL + +P E+Q+L++V AAALNP+D+KRR +D P
Sbjct: 286 KAWVYGEYGGVDVLKLDSNVTVPDVKEDQVLIKVVAAALNPVDAKRRQGKFKATDSPLPT 465
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
VPG D+AGVV+ G VK F +GDEVYG++ + ++E PKQ G+LA++ VEE L+A KP
Sbjct: 466 VPGYDVAGVVVKVGSQVKDFKVGDEVYGDVNE-KALEGPKQFGSLAEYTAVEEKLLAPKP 642
Query: 120 KRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
K L F +AASLPLA++TA EG + F G+++ V+ G+GGVG+LV+QLAK ++GAS V
Sbjct: 643 KNLDFAQAASLPLAIETAYEGLERTGFSPGKSILVLNGSGGVGSLVIQLAKQVYGASRVA 822
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDI 239
++ ST L +K GAD +DYTK +ED+ EKFD +YD +G C ++ K+DG++V +
Sbjct: 823 ATSSTRNLDLLKSLGADLAIDYTKENFEDLPEKFDVVYDAIGQCDRAVKAVKEDGSVVAL 1002
Query: 240 TWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRA 299
T + + +T G++L KL PYLE G++K ++DPKG + F+ + +AF Y+ET RA
Sbjct: 1003TGAVTPPGFRF-VVTSNGDVLRKLNPYLESGKVKPIVDPKGPFSFDKLAEAFSYLETNRA 1179
Query: 300 WGKVVVTCFP 309
GKVV+ P
Sbjct: 1180TGKVVIHPIP 1209
>TC226359 similar to UP|O23939 (O23939) Ripening-induced protein (Fragment),
partial (93%)
Length = 1355
Score = 285 bits (728), Expect = 3e-77
Identities = 153/311 (49%), Positives = 207/311 (66%), Gaps = 4/311 (1%)
Frame = +3
Query: 3 KAWFYEEYGP-KEVLKLGD-FPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FP 58
KAW Y EYG KEVLK +P E+Q+L++V AA+LNPID KR SD P
Sbjct: 159 KAWTYSEYGKSKEVLKFNQSVALPEVKEDQVLIKVAAASLNPIDHKRMEGYFKNSDSPLP 338
Query: 59 AVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARK 118
PG D+AGVV+ G VKKF +GDEVYG+I + ++E PK +G+LA++ EE L+A K
Sbjct: 339 TAPGYDVAGVVVKVGSEVKKFKVGDEVYGDI-NVKALEYPKVIGSLAEYTAAEERLLAHK 515
Query: 119 PKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
P+ LSF EAASLPL ++TA EG + F G+++ V+GGAGGVGT V+QLAK ++GAS V
Sbjct: 516 PQNLSFAEAASLPLTLETAYEGLERTGFSAGKSILVLGGAGGVGTHVIQLAKHVYGASKV 695
Query: 179 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVD 238
++ ST KL+ ++ GAD +DYTK +ED+ EKFD +YDTVG +++F V K+ G +V
Sbjct: 696 AATASTRKLELLRNLGADWPIDYTKENFEDLSEKFDVVYDTVGQTEQAFKVLKEGGKVVT 875
Query: 239 ITWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGR 298
I P H A+ L+ G ILEKL PY E G+LK ++DPK + F ++AF ++ET R
Sbjct: 876 IVPPGFHP-AILFILSTDGAILEKLNPYFESGKLKPILDPKSPFPFSQTVEAFAHLETNR 1052
Query: 299 AWGKVVVTCFP 309
A GK+VV P
Sbjct: 1053ATGKIVVYPIP 1085
>TC206726 similar to UP|O23939 (O23939) Ripening-induced protein (Fragment),
partial (84%)
Length = 1627
Score = 274 bits (700), Expect = 5e-74
Identities = 146/312 (46%), Positives = 207/312 (65%), Gaps = 5/312 (1%)
Frame = +3
Query: 3 KAWFYEEYG-PKEVLKLG-DFPIPSPL-ENQLLVQVYAAALNPIDSKRRMRPIFPSD--F 57
KAW+Y E+G P +VLKL ++P+P L ++Q+L++V AA++NP+D KR +D
Sbjct: 108 KAWYYSEHGSPGDVLKLDPNWPLPQQLKDDQVLIKVIAASINPVDYKRMHGEFKDTDPHL 287
Query: 58 PAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVAR 117
P VPG D+AG+V+ G VKKF +GDEVYG+I + + K GTL+++ + EE L+A
Sbjct: 288 PIVPGYDVAGIVVKVGGEVKKFKVGDEVYGDINE-QGLSNLKIHGTLSEYTIAEERLLAH 464
Query: 118 KPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASY 177
KP LSF EAAS+PLA++TA EGF+ F G+++ V+GGAGGVG V+QLAK ++ AS
Sbjct: 465 KPSNLSFIEAASIPLALETANEGFEHAHFSAGKSILVLGGAGGVGNYVIQLAKQVYKASK 644
Query: 178 VVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIV 237
+ ++ ST KL+ +++ G D +DYTK +ED+ EK+D +YD VG ++F K+DG +V
Sbjct: 645 IAATASTGKLELLRELGVDLPIDYTKENFEDLPEKYDLVYDVVGQGDRAFKAVKEDGKVV 824
Query: 238 DITWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETG 297
I P H A++ LT G ILE LRPY E G+LK ++D K F VI+A Y+ET
Sbjct: 825 TIV-PPGHPPAMFFVLTSKGSILENLRPYFESGKLKPILDAKTPVPFSQVIEAISYLETS 1001
Query: 298 RAWGKVVVTCFP 309
RA GKVVV P
Sbjct: 1002RATGKVVVYPIP 1037
>TC223408 similar to UP|Q43677 (Q43677) Auxin-induced protein, partial (54%)
Length = 543
Score = 162 bits (409), Expect = 3e-40
Identities = 85/175 (48%), Positives = 116/175 (65%), Gaps = 2/175 (1%)
Frame = +1
Query: 37 AAALNPIDSKRRMRPIFPSD--FPAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNS 94
A LNP D R + +D P VPG D AGVV+ G V KF +GDEVYG+I ++ +
Sbjct: 4 ARGLNPADYMRALGFFKDTDAPLPIVPGFDAAGVVVRVGSKVSKFKVGDEVYGDIIEY-A 180
Query: 95 MEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFV 154
PK +GTLA++ EE ++A KP LSF EAASLP A+ TA +GF +F G+++ V
Sbjct: 181 WNNPKTIGTLAEYTATEEKVLAHKPSNLSFIEAASLPAAIITAYQGFDKIEFSAGKSILV 360
Query: 155 VGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDI 209
+GGAGGVG+LV+QLAK +FGAS V ++ STPK ++ GAD +DYTK +E++
Sbjct: 361 LGGAGGVGSLVIQLAKHVFGASKVAATASTPKQDLLRSLGADLAIDYTKENFEEL 525
>TC226692 similar to UP|Q9AYU1 (Q9AYU1) Quinone-oxidoreductase QR1
(Fragment), partial (90%)
Length = 1229
Score = 138 bits (348), Expect = 3e-33
Identities = 106/335 (31%), Positives = 166/335 (48%), Gaps = 30/335 (8%)
Frame = +1
Query: 1 MQKAWFYEEYGPKEV-LKLGDFPIPSPLENQLLVQVYAAALNPID---SKRRMRPIF-PS 55
+ +A Y+ YG LK + +PSP N++L+++ A ++NPID K +RP+F P
Sbjct: 46 LMQAVRYDAYGGGPAGLKHVEVAVPSPKANEVLIKLEAVSINPIDWKIQKGLLRPLFLPR 225
Query: 56 DFPAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLV 115
FP +P D+AG ++ G VK F +GD+V + + G A+F V E+L
Sbjct: 226 TFPHIPCTDVAGEIVEIGTQVKDFKVGDQVLAKLTH-------QYGGGFAEFAVASESLT 384
Query: 116 ARKPKRLSFEEAASLPLAVQTA------IEGFKTGDFKKGETMFVVGGAGGVGTLVLQLA 169
A +P +S EAA+LP+A TA I G K + + + V +GGVG +QLA
Sbjct: 385 AARPSEVSAAEAAALPIAGLTARDALTQIAGVKLDGTGQLKNILVTAASGGVGHYAVQLA 564
Query: 170 KLMFGASYVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVV 229
KL G ++V ++C + FVK GAD+V+DY + ++ YD V +C
Sbjct: 565 KL--GNTHVTATCGARNIDFVKGLGADEVLDYRTPEGAALKSPSGRKYDAVINCTTGISW 738
Query: 230 AKKD------GAIVDIT------WPASHERAVYSS-------LTVCGEILEKLRPYLERG 270
+ D +VD+T W A+ ++ +S + V E LE L ++ G
Sbjct: 739 STFDPNLTEKAVVVDLTPNASSLWTAAMKKITFSKKQLVPFFVNVQREGLEYLLQLVKDG 918
Query: 271 ELKAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
+LK+VID K + DA+ G A GK++V
Sbjct: 919 KLKSVIDSK--FPLSKAEDAWAKSIDGHATGKIIV 1017
>AW397551 similar to PIR|T10824|T10 auxin-induced protein (clone MII-3) -
mung bean, partial (34%)
Length = 421
Score = 99.4 bits (246), Expect = 2e-21
Identities = 48/112 (42%), Positives = 71/112 (62%), Gaps = 2/112 (1%)
Frame = +1
Query: 200 DYTKTKYEDIEEKFDFIYDTVGDCK--KSFVVAKKDGAIVDITWPASHERAVYSSLTVCG 257
DYTK +E++EEKFD +YDTVG+ + K+ K+ G +V I H A++ G
Sbjct: 1 DYTKVNFEELEEKFDVVYDTVGESETEKALKAVKESGKVVTIV-RFGHPEAIFFIRISDG 177
Query: 258 EILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRAWGKVVVTCFP 309
+LEKL+PYLE G++K ++DPK Y F ++AF +++T RA GKVV+ P
Sbjct: 178 TVLEKLKPYLESGKVKPILDPKSPYPFSQTVEAFAHLKTNRAIGKVVIHPIP 333
>TC212554 similar to UP|Q43677 (Q43677) Auxin-induced protein, partial (30%)
Length = 439
Score = 87.4 bits (215), Expect = 8e-18
Identities = 42/96 (43%), Positives = 63/96 (64%)
Frame = +1
Query: 214 DFIYDTVGDCKKSFVVAKKDGAIVDITWPASHERAVYSSLTVCGEILEKLRPYLERGELK 273
D +YDTVG+ K+ K+ G +V I PA+ +S+++ G +LEKL+PYLE G++K
Sbjct: 4 DVVYDTVGESNKALKAVKEGGKVVTIVPPATPPAITFSAVSD-GAVLEKLQPYLESGKVK 180
Query: 274 AVIDPKGEYDFENVIDAFGYIETGRAWGKVVVTCFP 309
V+DPKG + F ++AF Y++T RA GKVV+ P
Sbjct: 181 PVLDPKGPFPFSQTVEAFAYLKTNRAIGKVVLHPIP 288
>TC218457 similar to PIR|T05166|T05166 quinone reductase homolog F18E5.200 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(97%)
Length = 1052
Score = 82.4 bits (202), Expect = 3e-16
Identities = 68/220 (30%), Positives = 104/220 (46%), Gaps = 9/220 (4%)
Frame = +2
Query: 11 GPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVP--GCDMAGV 68
G EVL+L + P +++LL+ V+A +LN D+ +R + +P A P G + +G
Sbjct: 77 GGPEVLQLEEVEDPQVGDDELLIGVHATSLNRADTFQR-KGSYPPPKGASPYLGLECSGT 253
Query: 69 VIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAA 128
V+ G NV + IGD+V + G A+ + V V P +S +AA
Sbjct: 254 VLSLGKNVSSWKIGDQVCALLAG----------GGYAEKVAVPVGQVLPVPAGVSLTDAA 403
Query: 129 SLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKL 187
S P T F +GET+ V GG+ G+GT +Q+AK G+ V++ S KL
Sbjct: 404 SFPEVACTVWSTVFMMSRLSQGETLLVHGGSSGIGTFAIQIAKYR-GSRVFVTAGSEEKL 580
Query: 188 KFVKQFGADKVVDY------TKTKYEDIEEKFDFIYDTVG 221
F K GAD ++Y + K E + D I D +G
Sbjct: 581 AFCKSIGADVGINYKTEDFVARVKEETGGQGVDVILDCMG 700
>TC205897 similar to UP|Q9SXZ7 (Q9SXZ7) NADPH oxidoreductase homolog
(Fragment), partial (44%)
Length = 1107
Score = 70.9 bits (172), Expect = 8e-13
Identities = 61/202 (30%), Positives = 91/202 (44%), Gaps = 34/202 (16%)
Frame = +3
Query: 142 KTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCST-PKLKFVKQFGADKVVD 200
K G + G+ + V AGG G +QLAKL + VV++C K K +K G ++V+D
Sbjct: 150 KAGQMESGKVVLVTAAAGGTGQFAVQLAKL--AGNTVVATCGGGAKAKLLKDLGVNRVID 323
Query: 201 Y----TKTKY-EDIEEKFDFIYDTVGD-----CKKSFVVAKKDGAIVDIT---------- 240
Y KT E+ + D IY++VG C + V + I I+
Sbjct: 324 YHSEDVKTVLREEFPKGIDIIYESVGGDMLNLCLNALAVHGRLIVIGMISQYQGEKGWTP 503
Query: 241 --WPASHERAVYSSLTVCG-----------EILEKLRPYLERGELKAVIDPKGEYDFENV 287
+P E+ + S TV G E L++L G+LK IDPK +
Sbjct: 504 SKYPGLLEKLLAKSQTVSGFFLVQYGHLWQEHLDRLFNLYSSGKLKVAIDPKKFIGLHSA 683
Query: 288 IDAFGYIETGRAWGKVVVTCFP 309
DA Y+ +G++ GKVVV+ P
Sbjct: 684 ADAVEYLHSGKSVGKVVVSVDP 749
>TC226693 weakly similar to UP|Q6T7C8 (Q6T7C8) Fiber quinone-oxidoreductase
(Fragment), partial (33%)
Length = 453
Score = 53.9 bits (128), Expect(2) = 9e-10
Identities = 30/80 (37%), Positives = 45/80 (55%)
Frame = +1
Query: 58 PAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVAR 117
P +P D+AG ++ G VK F +GD+V + + G LA+F+V E+L A
Sbjct: 226 PHIPCTDVAGEIVEIGPQVKDFKVGDKVLAKLNH-------QYGGGLAEFVVASESLTAA 384
Query: 118 KPKRLSFEEAASLPLAVQTA 137
+P +S EAA+LP+A TA
Sbjct: 385 RPSEVSAAEAAALPIAGLTA 444
Score = 26.6 bits (57), Expect(2) = 9e-10
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 3/34 (8%)
Frame = +2
Query: 23 IPSPLENQLLVQVYAAALNPID---SKRRMRPIF 53
+PSP N++L+++ +NPI K +RP+F
Sbjct: 122 VPSPKANEVLLKLEPICINPIHWKIQKGLLRPLF 223
>TC204440 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (97%)
Length = 1481
Score = 60.1 bits (144), Expect = 1e-09
Identities = 55/236 (23%), Positives = 106/236 (44%), Gaps = 27/236 (11%)
Frame = +3
Query: 12 PKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVIG 71
P +L + + + + + ++V+ + D + + S++P VPG ++ G V+
Sbjct: 111 PSGILSPYTYNLRNTGPDDVYIKVHYCGICHSDLHQIKNDLGMSNYPMVPGHEVVGEVLE 290
Query: 72 KGVNVKKFDIGDEV---------------YGNIQDFNSME-----------KPKQLGTLA 105
G +V +F +G+ V +I+++ S + KP Q G A
Sbjct: 291 VGSDVSRFRVGELVGVGLLVGCCKNCQPCQQDIENYCSKKIWSYNDVYVDGKPTQ-GGFA 467
Query: 106 QFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLV 165
+ ++VE+ V + P+ L+ E+ A L A T K+ + G GGVG +
Sbjct: 468 ETMIVEQKFVVKIPEGLAPEQVAPLLCAGVTVYSPLVHFGLKESGLRGGILGLGGVGHMG 647
Query: 166 LQLAKLMFGASYVVSSCSTPKLKFVKQFGADK-VVDYTKTKYEDIEEKFDFIYDTV 220
+++AK + V+SS K + ++ GAD+ +V T ++ + D+I DTV
Sbjct: 648 VKIAKALGHHVTVISSSDKKKQEALEHLGADQYLVSSDATAMQEAADSLDYIIDTV 815
>TC218715 similar to UP|Q84N44 (Q84N44) Oxidoreductase, partial (57%)
Length = 897
Score = 47.8 bits (112), Expect = 7e-06
Identities = 30/80 (37%), Positives = 44/80 (54%), Gaps = 1/80 (1%)
Frame = +3
Query: 143 TGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADKVVDYT 202
T +G+ + VVGG G VG +QLA + G S V ++C + + + GAD+ VDY
Sbjct: 33 TARISEGQRILVVGGGGAVGLSAVQLA-VAAGCS-VATTCGSQSVDRLLAAGADQAVDYV 206
Query: 203 KTKYE-DIEEKFDFIYDTVG 221
E I+ KFD + DT+G
Sbjct: 207 AEDVELAIKGKFDAVLDTIG 266
>TC211270 similar to UP|Q84N44 (Q84N44) Oxidoreductase, partial (29%)
Length = 567
Score = 45.8 bits (107), Expect = 3e-05
Identities = 27/89 (30%), Positives = 47/89 (52%), Gaps = 4/89 (4%)
Frame = +3
Query: 3 KAWFYEEYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRM---RPIFPSDFP 58
+A +G VL+L P+P + +LV+ A ++NP+D++ R R IF P
Sbjct: 255 RAVLLPSFGGPHVLQLRSHVPVPPLKPHDVLVRARAVSVNPLDTRMRAGYGRSIFEPLLP 434
Query: 59 AVPGCDMAGVVIGKGVNVKKFDIGDEVYG 87
+ G D++G V G V+ +G++V+G
Sbjct: 435 LILGRDVSGEVSAVGDKVRSVSVGEQVFG 521
>TC205898 similar to UP|Q9SXZ7 (Q9SXZ7) NADPH oxidoreductase homolog
(Fragment), partial (53%)
Length = 822
Score = 43.5 bits (101), Expect = 1e-04
Identities = 42/139 (30%), Positives = 63/139 (45%), Gaps = 7/139 (5%)
Frame = +1
Query: 102 GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGV 161
G A+F+++ P R E A L + +I K G + G+ + V AGG
Sbjct: 346 GGYAEFLMIPSKHALPVP-RPDPEVVAMLTSGLTASIALEKAGQMESGKVVLVTAAAGGT 522
Query: 162 GTLVLQLAKLMFGASYVVSSC-STPKLKFVKQFGADKVV----DYTKTKY-EDIEEKFDF 215
G +QLAK+ V++C K K +K G V+ D KT E++ + D
Sbjct: 523 GQFAVQLAKIP*STG--VATCGGGAKPKLLKDLGVHIVIDDHSDDVKTGLREELPKCIDV 696
Query: 216 IYDTV-GDCKKSFVVAKKD 233
IYD+V GD S + A+ D
Sbjct: 697 IYDSVGGDTINSCLSAEHD 753
>AI966462
Length = 470
Score = 43.5 bits (101), Expect = 1e-04
Identities = 37/114 (32%), Positives = 47/114 (40%), Gaps = 18/114 (15%)
Frame = +3
Query: 126 EAASLPLAVQTAIEG----------FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGA 175
E +PL+ T I G F+ G KKGE +FV +G VG LV Q AKL
Sbjct: 36 EHTDVPLSYYTGILGMPGMTAYAGFFELGSPKKGENVFVSAASGAVGQLVGQFAKL--AG 209
Query: 176 SYVVSSCSTP--------KLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVG 221
YVV S + KL F + F + D T E D ++ VG
Sbjct: 210 CYVVGSAGSKEKVDLLKNKLGFDEAFNYKEESDLNTTLKSYFPEGIDIYFENVG 371
>TC204951 similar to UP|Q8LEB8 (Q8LEB8) Quinone oxidoreductase-like protein,
partial (14%)
Length = 440
Score = 40.8 bits (94), Expect(2) = 3e-04
Identities = 20/40 (50%), Positives = 30/40 (75%), Gaps = 1/40 (2%)
Frame = +3
Query: 10 YGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRR 48
+G +VLKL + +P E+Q+L++V AAALNP+D+KRR
Sbjct: 306 WGGXDVLKLDSNVAVPDVKEDQVLIKVVAAALNPVDAKRR 425
Score = 20.8 bits (42), Expect(2) = 3e-04
Identities = 7/9 (77%), Positives = 7/9 (77%)
Frame = +1
Query: 3 KAWFYEEYG 11
KAW Y EYG
Sbjct: 283 KAWVYGEYG 309
>TC204521 similar to UP|Q9SLN8 (Q9SLN8) Allyl alcohol dehydrogenase, partial
(94%)
Length = 1230
Score = 42.4 bits (98), Expect = 3e-04
Identities = 36/114 (31%), Positives = 47/114 (40%), Gaps = 18/114 (15%)
Frame = +2
Query: 126 EAASLPLAVQTAIEG----------FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGA 175
E +PL+ T I G F+ G KKG+T+FV +G VG LV Q AKL
Sbjct: 392 EHTDVPLSYYTGILGMPGMTAYAGFFEVGSPKKGDTVFVSAASGAVGQLVGQFAKLT--G 565
Query: 176 SYVVSSCSTP--------KLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVG 221
YVV S + KL F + F + D E D ++ VG
Sbjct: 566 CYVVGSAGSKEKVDLLKNKLGFDEAFNYKEEPDLNAALKRYFPEGIDIYFENVG 727
>AW508216 similar to PIR|T49047|T49 quinone reductase-like protein -
Arabidopsis thaliana, partial (37%)
Length = 423
Score = 41.2 bits (95), Expect = 7e-04
Identities = 27/79 (34%), Positives = 38/79 (47%)
Frame = +2
Query: 58 PAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVAR 117
P +PG D +G V G V KF +GD V LG+ AQFIVV+E+ + +
Sbjct: 215 PFIPGSDFSGFVDAVGSKVSKFRVGDAVCSF----------AGLGSFAQFIVVDESQLFQ 364
Query: 118 KPKRLSFEEAASLPLAVQT 136
P+ A +L +A T
Sbjct: 365 VPQGCDLVAAGALAVASGT 421
>TC210667 similar to UP|Q6WAU0 (Q6WAU0) (+)-pulegone reductase, partial (52%)
Length = 539
Score = 40.8 bits (94), Expect = 9e-04
Identities = 31/89 (34%), Positives = 39/89 (42%), Gaps = 8/89 (8%)
Frame = +3
Query: 141 FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTP--------KLKFVKQ 192
F+ G KKGE +FV +G VG LV Q AKL YVV S + KL F +
Sbjct: 3 FEVGSPKKGENVFVSAASGAVGQLVGQFAKLT--DCYVVGSAGSKEKVDLLKNKLGFDEA 176
Query: 193 FGADKVVDYTKTKYEDIEEKFDFIYDTVG 221
F + D T E D ++ VG
Sbjct: 177 FNYKEESDLNATLKRYFPEGIDIYFENVG 263
>TC206457 weakly similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol
dehydrogenase (NAD-dependent mannitol dehydrogenase) ,
partial (75%)
Length = 1180
Score = 40.4 bits (93), Expect = 0.001
Identities = 39/123 (31%), Positives = 53/123 (42%), Gaps = 4/123 (3%)
Frame = +3
Query: 102 GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDF--KKGETMFVVGGAG 159
G +Q V + V P+ L+ + AA L A T K D G+ + VVG G
Sbjct: 222 GGYSQIFVADYRYVVHIPENLAMDAAAPLLCAGITVFNPLKDHDLVASPGKKIGVVG-LG 398
Query: 160 GVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQ-FGADK-VVDYTKTKYEDIEEKFDFIY 217
G+G + ++ K FG V S S K KQ GAD +V + + DFI
Sbjct: 399 GLGHIAVKFGK-AFGHHVTVISTSPSKEAEAKQRLGADDFIVSSNPKQLQAARRSIDFIL 575
Query: 218 DTV 220
DTV
Sbjct: 576 DTV 584
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.139 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,119,195
Number of Sequences: 63676
Number of extensions: 158379
Number of successful extensions: 720
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 694
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 701
length of query: 340
length of database: 12,639,632
effective HSP length: 98
effective length of query: 242
effective length of database: 6,399,384
effective search space: 1548650928
effective search space used: 1548650928
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146342.2


