

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.6 + phase: 0
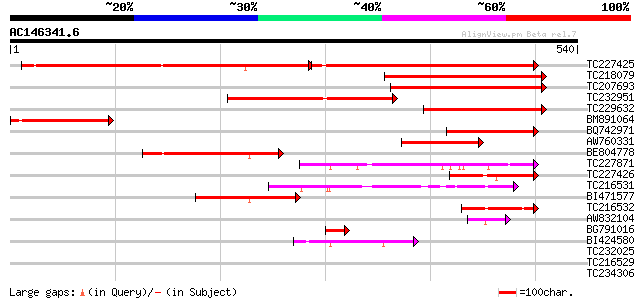
(540 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227425 similar to UP|Q8HWA9 (Q8HWA9) Mus musculus 0 day neonat... 262 e-133
TC218079 similar to UP|Q945B9 (Q945B9) Growth-on protein GRO11, ... 294 8e-80
TC207693 similar to UP|Q945B9 (Q945B9) Growth-on protein GRO11, ... 282 2e-76
TC232951 similar to UP|Q945B8 (Q945B8) Growth-on protein GRO10, ... 225 4e-59
TC229632 similar to UP|Q945B8 (Q945B8) Growth-on protein GRO10, ... 204 6e-53
BM891064 similar to PIR|D86147|D861 T1N6.3 protein - Arabidopsis... 174 1e-43
BQ742971 165 5e-41
AW760331 similar to GP|16118856|gb| growth-on protein GRO10 {Eup... 115 4e-26
BE804778 103 3e-22
TC227871 similar to UP|Q93Z32 (Q93Z32) AT4g33410/F17M5_170, part... 97 1e-20
TC227426 similar to UP|Q8LQG1 (Q8LQG1) Vacuolar sorting receptor... 93 3e-19
TC216531 similar to UP|Q9VPQ7 (Q9VPQ7) CG11840-PA (SD07518p), pa... 85 9e-17
BI471577 83 4e-16
TC216532 similar to UP|Q6SXP6 (Q6SXP6) Signal peptide peptidase ... 62 8e-10
AW832104 similar to GP|16118854|gb| growth-on protein GRO11 {Eup... 60 2e-09
BG791016 weakly similar to GP|16118856|gb| growth-on protein GRO... 44 1e-04
BI424580 similar to GP|16648809|gb AT4g33410/F17M5_170 {Arabidop... 44 2e-04
TC232025 similar to UP|P93484 (P93484) BP-80 vacuolar sorting re... 39 0.008
TC216529 similar to UP|Q6SXP6 (Q6SXP6) Signal peptide peptidase ... 38 0.010
TC234306 homologue to UP|Q93X09 (Q93X09) Vacuolar sorting recept... 33 0.25
>TC227425 similar to UP|Q8HWA9 (Q8HWA9) Mus musculus 0 day neonate cerebellum
cDNA, RIKEN full-length enriched library,
clone:C230098O20 product:histocompatibility 13, full
insert sequence, partial (6%)
Length = 2073
Score = 262 bits (669), Expect(2) = e-133
Identities = 124/216 (57%), Positives = 159/216 (73%)
Frame = +3
Query: 288 EGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYA 347
EG+ + +L R Q Q V +P FG + +LAV FC+ FA+ WA RQ SY+
Sbjct: 1008 EGMHNWIVSLT--LRKCQNCGQKTVSLPLFGEISIFSLAVLLFCVAFAIFWAATRQESYS 1181
Query: 348 WIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARG 407
WIGQDILGI L+ITVLQ+ R+PN+KV TVLL CAF+YDI WVF+S FHESVMI VARG
Sbjct: 1182 WIGQDILGICLMITVLQLARLPNIKVATVLLCCAFVYDIFWVFISPVIFHESVMIAVARG 1361
Query: 408 DKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVW 467
DK+G + IPMLL+ PRLFDPWGGY +IGFGDI+ PGL+++F+ R+D + GYF+W
Sbjct: 1362 DKAGGEAIPMLLRFPRLFDPWGGYDMIGFGDILFPGLLISFAHRFDKDNGRGASNGYFLW 1541
Query: 468 AMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTLG 503
+ YG+GL++TY+ L LM+G+GQPALLY+VP TLG
Sbjct: 1542 LVVGYGIGLVLTYLGLYLMNGNGQPALLYLVPCTLG 1649
Score = 230 bits (587), Expect(2) = e-133
Identities = 126/280 (45%), Positives = 168/280 (60%), Gaps = 2/280 (0%)
Frame = +2
Query: 12 VAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGP 71
V V LF + A D DD AP C N F LVKV W+DG E Y GV ARFG
Sbjct: 179 VLLVLLFVIGAG-ADDAKRDDDRAPKSESCNNPFQLVKVENWVDGEEGHIYNGVSARFGS 355
Query: 72 TLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAIL 131
L K ++ T + ADP DCCS ++L+ + L RG C FT KA+ A GA+A+L
Sbjct: 356 VLPEKPDNSVKTPAIFADPLDCCSNSTSRLSGSVALCVRGGCDFTVKADFAQSVGATAML 535
Query: 132 IINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVD 191
+IN +LF+MVC N T+ +I IP VM+ + AG +L + + + S V I LY+P RPLVD
Sbjct: 536 VINDAQDLFEMVC-SNSTEANISIPVVMITKSAGQSLNKSLTSGSKVEILLYAPPRPLVD 712
Query: 192 VAEVFLWLMAVGTILCASYWSAWTAREAAIEQ--EKLLKDASDEYVAESVGSRGYVEIST 249
+ FLWLM++GTI+CAS WS T E + E+ E K++S+ A+ V I +
Sbjct: 713 FSVAFLWLMSIGTIVCASLWSDLTTPEKSDERYNELCPKESSNADTAKDDSDNEIVNIDS 892
Query: 250 TAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEG 289
A++FV+ AS FLV+L+ MS WF+ VL+VLFCIGGI G
Sbjct: 893 KGAVIFVIAASTFLVLLFFFMSSWFVWVLIVLFCIGGIRG 1012
>TC218079 similar to UP|Q945B9 (Q945B9) Growth-on protein GRO11, partial
(33%)
Length = 1909
Score = 294 bits (752), Expect = 8e-80
Identities = 141/154 (91%), Positives = 147/154 (94%)
Frame = +2
Query: 358 LIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPM 417
LIITVLQIVRIPNLKVGTVLLSCAFLYDI WVFVSKWWFHESVMIVVARGD+SGEDGIPM
Sbjct: 2 LIITVLQIVRIPNLKVGTVLLSCAFLYDIFWVFVSKWWFHESVMIVVARGDRSGEDGIPM 181
Query: 418 LLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLL 477
LLK+PR+FDPWGGYSIIGFGDIILPGL+VAFSLRYDWLAKKNLR GYF+WAMTAYGLGLL
Sbjct: 182 LLKIPRMFDPWGGYSIIGFGDIILPGLLVAFSLRYDWLAKKNLRDGYFLWAMTAYGLGLL 361
Query: 478 ITYVALNLMDGHGQPALLYIVPFTLGKYNHLTHK 511
ITYVALNLMDGHGQPALLYIVPFTLG + L K
Sbjct: 362 ITYVALNLMDGHGQPALLYIVPFTLGTFLSLGKK 463
>TC207693 similar to UP|Q945B9 (Q945B9) Growth-on protein GRO11, partial
(32%)
Length = 1071
Score = 282 bits (722), Expect = 2e-76
Identities = 136/149 (91%), Positives = 141/149 (94%)
Frame = +3
Query: 363 LQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLP 422
LQIV +PNLKVGTVLLSCAFLYDI WVFVSK WFHESVMIVVARGDKSGEDGIPMLLK+P
Sbjct: 3 LQIVSLPNLKVGTVLLSCAFLYDIFWVFVSKRWFHESVMIVVARGDKSGEDGIPMLLKIP 182
Query: 423 RLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVA 482
RLFDPWGGYSIIGFGDIILPGL+VAFSLRYDWLAKKNLRAGYF+WAMTAYGLGLLITYVA
Sbjct: 183 RLFDPWGGYSIIGFGDIILPGLIVAFSLRYDWLAKKNLRAGYFLWAMTAYGLGLLITYVA 362
Query: 483 LNLMDGHGQPALLYIVPFTLGKYNHLTHK 511
LNLMDGHGQPALLYIVPFTLG + L K
Sbjct: 363 LNLMDGHGQPALLYIVPFTLGTFLSLGKK 449
>TC232951 similar to UP|Q945B8 (Q945B8) Growth-on protein GRO10, partial
(29%)
Length = 502
Score = 225 bits (573), Expect = 4e-59
Identities = 113/163 (69%), Positives = 132/163 (80%), Gaps = 1/163 (0%)
Frame = +1
Query: 208 ASYWSAWTAREAAIEQEKLLKDASDEYVAESVGS-RGYVEISTTAAILFVVLASCFLVML 266
A+ SAWT REAAIEQ+KLLKDASDE S G V ++ AA+LFVV ASCFL ML
Sbjct: 19 AARGSAWTTREAAIEQDKLLKDASDELPNTKYASVSGVVNMNVKAAVLFVVFASCFLFML 198
Query: 267 YKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPYLTLA 326
YKLMS WF++VLVVLFCIGGIEGLQTCL ALLS RWF++ ++Y+K+PF GA+ YLTLA
Sbjct: 199 YKLMSSWFIDVLVVLFCIGGIEGLQTCLVALLS--RWFKHAGESYIKVPFLGAISYLTLA 372
Query: 327 VTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIP 369
V+PFCI FA++WAV R S+AWIGQDILGIALIITVLQIV +P
Sbjct: 373 VSPFCITFAILWAVYRNESFAWIGQDILGIALIITVLQIVHVP 501
>TC229632 similar to UP|Q945B8 (Q945B8) Growth-on protein GRO10, partial
(25%)
Length = 620
Score = 204 bits (520), Expect = 6e-53
Identities = 96/117 (82%), Positives = 106/117 (90%)
Frame = +2
Query: 395 WFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDW 454
+F ESVMIVVARGD+SGEDGIPMLLK PR+FDPWGGYSIIGFGDI+LPG++VAFSLRYDW
Sbjct: 2 FFKESVMIVVARGDRSGEDGIPMLLKFPRIFDPWGGYSIIGFGDILLPGMLVAFSLRYDW 181
Query: 455 LAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTLGKYNHLTHK 511
LA K+LR+GYF+WAM AYG GLL+TYVALNLMDGHGQPALLYIVPFTLG L K
Sbjct: 182 LANKSLRSGYFLWAMFAYGFGLLVTYVALNLMDGHGQPALLYIVPFTLGTLMTLGRK 352
>BM891064 similar to PIR|D86147|D861 T1N6.3 protein - Arabidopsis thaliana,
partial (12%)
Length = 421
Score = 174 bits (440), Expect = 1e-43
Identities = 86/100 (86%), Positives = 91/100 (91%), Gaps = 1/100 (1%)
Frame = +1
Query: 1 MVSLGVIYSNIVAYVFLF-SVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVEN 59
MVSLG IYS V YVFLF +V+LVL GDIVHHDDVAP RPGCENNFVLVKVPTWIDGVEN
Sbjct: 124 MVSLGAIYS--VIYVFLFGAVTLVLGGDIVHHDDVAPRRPGCENNFVLVKVPTWIDGVEN 297
Query: 60 AEYVGVGARFGPTLESKEKHANHTRVVMADPPDCCSKPKN 99
+EYVGVGARFGPTLESKEK AN +RVVMADPPDCC+KPKN
Sbjct: 298 SEYVGVGARFGPTLESKEKRANLSRVVMADPPDCCTKPKN 417
>BQ742971
Length = 423
Score = 165 bits (417), Expect = 5e-41
Identities = 73/87 (83%), Positives = 83/87 (94%)
Frame = +2
Query: 417 MLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGL 476
++LK PR+FDPWGGYSIIGFGDI+LPG++VAFSLRYDWLA K+LR+GYF+WAM AYG GL
Sbjct: 128 LILKFPRIFDPWGGYSIIGFGDILLPGMLVAFSLRYDWLANKSLRSGYFLWAMFAYGFGL 307
Query: 477 LITYVALNLMDGHGQPALLYIVPFTLG 503
L+TYVALNLMDGHGQPALLYIVPFTLG
Sbjct: 308 LVTYVALNLMDGHGQPALLYIVPFTLG 388
>AW760331 similar to GP|16118856|gb| growth-on protein GRO10 {Euphorbia
esula}, partial (14%)
Length = 235
Score = 115 bits (289), Expect = 4e-26
Identities = 55/78 (70%), Positives = 64/78 (81%)
Frame = +2
Query: 374 GTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSI 433
GT L+ AF+YDI WV VSK +F ESVMIVVARGD+SG DGIPM LK PR+FDPWGGYSI
Sbjct: 2 GTGLVXWAFIYDIFWVLVSKKFFKESVMIVVARGDRSGXDGIPMWLKFPRIFDPWGGYSI 181
Query: 434 IGFGDIILPGLVVAFSLR 451
I GD++L ++VAFSLR
Sbjct: 182 IXLGDMLLAXMLVAFSLR 235
>BE804778
Length = 416
Score = 103 bits (256), Expect = 3e-22
Identities = 58/136 (42%), Positives = 85/136 (61%), Gaps = 2/136 (1%)
Frame = +1
Query: 127 ASAILIINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPL 186
A+AIL+IN +LF+MVC N ++ +I IP VM+ + AG +L + + S V I LY+P
Sbjct: 10 ATAILVINDSQDLFEMVCS-NSSEANISIPVVMIAKSAGQSLNKSFTSGSKVEILLYAPP 186
Query: 187 RPLVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLL--KDASDEYVAESVGSRGY 244
RPLVD + FLWLM+VGTI+CAS WS T E + E+ L K++S+ + +
Sbjct: 187 RPLVDFSVAFLWLMSVGTIVCASLWSDLTTPEKSDERYNKLCPKESSNAETEKDDLDKEI 366
Query: 245 VEISTTAAILFVVLAS 260
V I + A++FV+ AS
Sbjct: 367 VNIDSKGAVIFVIAAS 414
>TC227871 similar to UP|Q93Z32 (Q93Z32) AT4g33410/F17M5_170, partial (96%)
Length = 1436
Score = 97.4 bits (241), Expect = 1e-20
Identities = 80/276 (28%), Positives = 131/276 (46%), Gaps = 49/276 (17%)
Frame = +3
Query: 277 VLVVLFCIGGIEGLQTCLTALLSCFRWF--QYPAQTYVKIPFFGAVPYLTLAVTP----- 329
+L++ + + L T TA+ S F P Y+K F A P+++ +
Sbjct: 429 LLLMFYLFSSVSQLLTAFTAVASASSLFFCLSPYAAYLKAQFGLADPFVSRCCSKSFTRI 608
Query: 330 ------FCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFL 383
C W V S WI ++LGI++ + + VR+PN+K+ +LL C F+
Sbjct: 609 QAILLLVCSFTVAAWLV----SGHWILNNLLGISICVAFVSHVRLPNIKICAMLLLCLFV 776
Query: 384 YDILWVFVSKWWFHESVMIVVARGDKS-------GEDGIPML------LKLP-RLFDP-- 427
YDI WVF S+ +F +VM+ VA S G+P L L+LP ++ P
Sbjct: 777 YDIFWVFFSERFFGANVMVSVATQQASNPVHTVANSIGLPGLQLITKKLELPVKIVFPRN 956
Query: 428 -WGG---------YSIIGFGDIILPGLVVAFSLRYDW---------LAKKNLRAGYFVW- 467
GG + ++G GD+ +PG+++A L +D+ L + + ++W
Sbjct: 957 LLGGVVPGENAADFMMLGLGDMAIPGMLLALVLCFDYRKSRDTVNLLELHSSKGHKYIWY 1136
Query: 468 AMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTLG 503
A+ Y +G L+T +A ++ QPALLY+VP TLG
Sbjct: 1137ALPGYAIG-LVTALAAGVLTHSPQPALLYLVPSTLG 1241
>TC227426 similar to UP|Q8LQG1 (Q8LQG1) Vacuolar sorting receptor-like
protein, partial (9%)
Length = 714
Score = 92.8 bits (229), Expect = 3e-19
Identities = 44/87 (50%), Positives = 62/87 (70%), Gaps = 3/87 (3%)
Frame = +2
Query: 420 KLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRA---GYFVWAMTAYGLGL 476
+ PRLFD GY +IG GDI+ PGL+++++ R L K N R YF+W + YG+GL
Sbjct: 14 RFPRLFDXXXGYDMIGLGDILFPGLLISYAHR---LXKDNRRXXSNAYFLWLVVXYGIGL 184
Query: 477 LITYVALNLMDGHGQPALLYIVPFTLG 503
++TY+ L LM+G+GQPALLY+VP T+G
Sbjct: 185 VLTYMGLYLMNGNGQPALLYLVPCTVG 265
>TC216531 similar to UP|Q9VPQ7 (Q9VPQ7) CG11840-PA (SD07518p), partial (11%)
Length = 942
Score = 84.7 bits (208), Expect = 9e-17
Identities = 70/251 (27%), Positives = 117/251 (45%), Gaps = 13/251 (5%)
Frame = +3
Query: 247 ISTTAAILFVVLASCFLVMLYKLMSFWFLE----VLVVLFCIGGIEGLQTCLTALLSCF- 301
+S A+ F + S L+ L+ L F + VL F + GI L L + F
Sbjct: 33 MSNEHAMRFPFVGSAMLLSLFLLFKFLSKDLVNTVLTGYFFVLGIVALSATLLPFIKRFL 212
Query: 302 --RW------FQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDI 353
W +++P ++I F + + T FC +A+ W+ +I
Sbjct: 213 PKHWNEDLIVWRFPYFRSLEIEFTKSQVVAGIPGTFFCAWYAL--------QKHWLANNI 368
Query: 354 LGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGED 413
LG+A I ++++ + + K G +LL+ F+YDI WVF + VM+ VA+ +
Sbjct: 369 LGLAFCIQGIEMLSLGSFKTGAILLAGLFVYDIFWVFFT------PVMVSVAKSFDA--- 521
Query: 414 GIPMLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYG 473
P+ L P D S++G GDI++PG+ VA +LR+D + + YF A Y
Sbjct: 522 --PIKLLFPTA-DSARPISMLGLGDIVIPGIFVALALRFD--VSRGKQPQYFKSAFVGYT 686
Query: 474 LGLLITYVALN 484
+GL++T V +N
Sbjct: 687 VGLVLTIVVMN 719
>BI471577
Length = 421
Score = 82.8 bits (203), Expect = 4e-16
Identities = 45/102 (44%), Positives = 64/102 (62%), Gaps = 2/102 (1%)
Frame = +1
Query: 178 VSIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLL--KDASDEYV 235
+ I LY+P RPLVD + FLWLM+VGTI+CAS WS T E + E+ L K++S+
Sbjct: 115 LEILLYAPPRPLVDFSVAFLWLMSVGTIVCASLWSDLTTPEKSDERYNKLCPKESSNAET 294
Query: 236 AESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLEV 277
+ + V I + A++FV+ AS FLV+L+ MS WF+ V
Sbjct: 295 EKDDLDKEIVNIDSKGAVIFVIAASTFLVLLFFFMSTWFVWV 420
>TC216532 similar to UP|Q6SXP6 (Q6SXP6) Signal peptide peptidase (Fragment),
partial (80%)
Length = 553
Score = 61.6 bits (148), Expect = 8e-10
Identities = 31/73 (42%), Positives = 46/73 (62%)
Frame = +3
Query: 431 YSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHG 490
+S++G GDI++PG+ VA +LR+D + + YF A Y +GL++T V +N
Sbjct: 60 FSMLGLGDIVIPGIFVALALRFD--VSRGKQPQYFKSAFVGYTVGLVLTIVVMNWFQA-A 230
Query: 491 QPALLYIVPFTLG 503
QPALLYIVP +G
Sbjct: 231 QPALLYIVPSVIG 269
>AW832104 similar to GP|16118854|gb| growth-on protein GRO11 {Euphorbia
esula}, partial (7%)
Length = 336
Score = 60.1 bits (144), Expect = 2e-09
Identities = 37/88 (42%), Positives = 40/88 (45%), Gaps = 47/88 (53%)
Frame = +2
Query: 437 GDIILPGLVVAFSLR--------------------------------------------- 451
GDIILPGL+VAFSLR
Sbjct: 5 GDIILPGLLVAFSLRLNRKSYINDPIHHLVLLTKTFWIDEDNALCLVNGS*T*FF**LTI 184
Query: 452 --YDWLAKKNLRAGYFVWAMTAYGLGLL 477
YDWLAKKNLR GYF+WAMTAYGLG+L
Sbjct: 185 YRYDWLAKKNLRDGYFLWAMTAYGLGML 268
>BG791016 weakly similar to GP|16118856|gb| growth-on protein GRO10
{Euphorbia esula}, partial (4%)
Length = 384
Score = 44.3 bits (103), Expect = 1e-04
Identities = 17/23 (73%), Positives = 21/23 (90%)
Frame = +2
Query: 301 FRWFQYPAQTYVKIPFFGAVPYL 323
F+WFQ+ AQT+VK+PFFGAV YL
Sbjct: 314 FKWFQHAAQTFVKVPFFGAVSYL 382
>BI424580 similar to GP|16648809|gb AT4g33410/F17M5_170 {Arabidopsis
thaliana}, partial (29%)
Length = 381
Score = 43.9 bits (102), Expect = 2e-04
Identities = 38/128 (29%), Positives = 58/128 (44%), Gaps = 9/128 (7%)
Frame = +3
Query: 271 SFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWF--QYPAQTYVKIPFFGAVPYLTLAVT 328
SF F + LF + L T TA+ S F P Y+K F A P+++ +
Sbjct: 3 SFSFFPHRIYLF--SSVSQLLTAFTAVASASSLFFCLSPYVAYLKAQFGLADPFVSRCCS 176
Query: 329 P-FCIVFAVVWAVKRQASYAWIGQDIL------GIALIITVLQIVRIPNLKVGTVLLSCA 381
F + A++ V AW+ I++ I + VR+PN+K+ +LL C
Sbjct: 177 KSFTRLQAILLLVCSFTVAAWLVSGHCYSFPPPRISICIAFVSHVRLPNIKICAMLLVCL 356
Query: 382 FLYDILWV 389
F+YDI WV
Sbjct: 357 FVYDIFWV 380
>TC232025 similar to UP|P93484 (P93484) BP-80 vacuolar sorting receptor,
partial (31%)
Length = 832
Score = 38.5 bits (88), Expect = 0.008
Identities = 26/84 (30%), Positives = 43/84 (50%), Gaps = 6/84 (7%)
Frame = +1
Query: 105 IILVHRGKCSFTTKANIADEAGASAILII-NYRTELFKMVCEENETD-----VDIGIPAV 158
I+L+ RG C F K A +AGASA+L+ + +L M E + +I IP+
Sbjct: 457 IVLLDRGNCFFALKVWNAQKAGASAVLVSDDIEEKLITMDTPEEDGSSAKYIENITIPSA 636
Query: 159 MLPQDAGLNLERHIQNNSIVSIQL 182
++ + G L+ I N +V++ L
Sbjct: 637 LIEKSFGEKLKVAISNGDMVNVNL 708
>TC216529 similar to UP|Q6SXP6 (Q6SXP6) Signal peptide peptidase (Fragment),
partial (57%)
Length = 513
Score = 38.1 bits (87), Expect = 0.010
Identities = 21/55 (38%), Positives = 31/55 (56%)
Frame = +2
Query: 449 SLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTLG 503
+LR+D + + YF A Y +GL++T + +N QPALLYIVP +G
Sbjct: 5 ALRFD--VSRGKQPQYFKSAFLGYTVGLVLTIIVMNWFQA-AQPALLYIVPSVIG 160
>TC234306 homologue to UP|Q93X09 (Q93X09) Vacuolar sorting receptor, partial
(21%)
Length = 639
Score = 33.5 bits (75), Expect = 0.25
Identities = 20/44 (45%), Positives = 25/44 (56%)
Frame = +1
Query: 95 SKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTE 138
SKP T +LV RG C FT KA A GA+AIL+ + + E
Sbjct: 484 SKPGGSPT--FLLVDRGDCYFTLKAWNAQNGGAAAILVADDKAE 609
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.142 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,306,767
Number of Sequences: 63676
Number of extensions: 417347
Number of successful extensions: 2901
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 2863
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2889
length of query: 540
length of database: 12,639,632
effective HSP length: 102
effective length of query: 438
effective length of database: 6,144,680
effective search space: 2691369840
effective search space used: 2691369840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146341.6


