

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.5 + phase: 0 /pseudo
(688 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
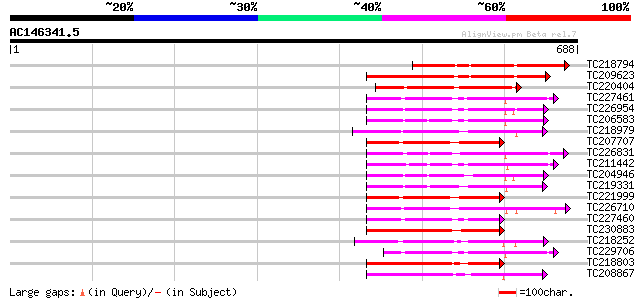
Score E
Sequences producing significant alignments: (bits) Value
TC218794 similar to PIR|T02456|T02456 protein kinase homolog F4I... 274 1e-73
TC209623 similar to UP|Q9FZP2 (Q9FZP2) Receptor-like protein kin... 178 6e-45
TC220404 weakly similar to PIR|T02456|T02456 protein kinase homo... 163 2e-40
TC227461 similar to UP|Q84SA6 (Q84SA6) Serine/threonine protein ... 147 2e-35
TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 144 2e-34
TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 135 5e-32
TC218979 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associate... 135 6e-32
TC207707 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase... 134 2e-31
TC226831 similar to PIR|T52285|T52285 serine/threonine-specific ... 133 2e-31
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 133 3e-31
TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 132 4e-31
TC219331 similar to UP|Q9FM85 (Q9FM85) Protein kinase-like prote... 132 7e-31
TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase... 132 7e-31
TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-l... 131 9e-31
TC227460 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protei... 131 9e-31
TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete 131 9e-31
TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial ... 131 9e-31
TC229706 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protei... 130 1e-30
TC218803 similar to UP|Q84P72 (Q84P72) Receptor-like protein kin... 130 2e-30
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 129 3e-30
>TC218794 similar to PIR|T02456|T02456 protein kinase homolog F4I18.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(30%)
Length = 998
Score = 274 bits (700), Expect = 1e-73
Identities = 136/190 (71%), Positives = 159/190 (83%)
Frame = +2
Query: 490 RIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNN 549
RIA ELCSALIFLHS+KPHS+VHGDLKPSNILLDANL++KLSDFGICR+LS N SS +
Sbjct: 2 RIAAELCSALIFLHSSKPHSVVHGDLKPSNILLDANLISKLSDFGICRILS--NCESSGS 175
Query: 550 STTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYA 609
+TT+FW T KGTF YMDPEFL +GELT KSDVYSFGI+LLRL+TG+PALGI EV YA
Sbjct: 176 NTTEFWRTD-PKGTFVYMDPEFLASGELTPKSDVYSFGIILLRLLTGRPALGITKEVKYA 352
Query: 610 LNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPMKVSC 669
L+ G +KS+LDPLAGDWP V+AE+L ALRCCDMN+KSRP+L S+ WRVL+ M+VS
Sbjct: 353 LDT--GKLKSLLDPLAGDWPFVQAEQLARLALRCCDMNRKSRPDLYSDVWRVLDAMRVSS 526
Query: 670 SGTNNFGLKS 679
G N+FGL S
Sbjct: 527 GGANSFGLSS 556
>TC209623 similar to UP|Q9FZP2 (Q9FZP2) Receptor-like protein kinase, partial
(33%)
Length = 986
Score = 178 bits (452), Expect = 6e-45
Identities = 92/225 (40%), Positives = 141/225 (61%), Gaps = 1/225 (0%)
Frame = +3
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHL-SRNGNNNAPPLTWQTRIRI 491
+V +L ++HPN++ LIG E L+YEY+ GSLED + + +W+ R I
Sbjct: 27 EVNILGCIRHPNMVLLIGACAEHGILVYEYMAKGSLEDCMFGKKKEKEGKMRSWKVRYGI 206
Query: 492 ATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNST 551
A E+ + L+FLH KP +VH DLKP NILLD N V+K+SD G+ +++ +++ +
Sbjct: 207 AAEIATGLLFLHQTKPEPLVHRDLKPGNILLDQNYVSKISDVGLAKLV----PAATAGNG 374
Query: 552 TQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALN 611
TQ +T+ A GTF Y+DPE+ TG L KSDVYS GI+LL+L+TG+PA+G+ ++V ++
Sbjct: 375 TQCCMTA-AAGTFCYIDPEYQQTGMLGVKSDVYSLGIILLQLLTGRPAMGLAHQVEESIK 551
Query: 612 NAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCS 656
+LDP DWP+ +A L + AL+C + +K RP+L +
Sbjct: 552 K--DRFGEMLDPSVPDWPLEQALCLANLALQCAQLRRKDRPDLAT 680
>TC220404 weakly similar to PIR|T02456|T02456 protein kinase homolog F4I18.11
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(11%)
Length = 510
Score = 163 bits (413), Expect = 2e-40
Identities = 84/178 (47%), Positives = 123/178 (68%)
Frame = +3
Query: 444 NLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIATELCSALIFLH 503
+LITL+GV E+ +++YEYLPNG+L+D+L R NN+ PLTW TR R+ E+ SAL FLH
Sbjct: 3 HLITLLGVCPEAWSIVYEYLPNGTLQDYLFRKSNNS--PLTWNTRARMIAEIASALCFLH 176
Query: 504 SNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGT 563
S KP +I+HGDLKP +LLD++L K+ FG+CR++ S + F +++ KG
Sbjct: 177 SFKPETIIHGDLKPETVLLDSSLGCKMCGFGLCRLV-----SEESLLRPSFRLSTEPKGA 341
Query: 564 FAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNNAGGNVKSVL 621
F Y DPEF TG LT+KSD+YSFG+++L+L+TG+ +G+ V A+ + G + S+L
Sbjct: 342 FTYTDPEFQRTGILTTKSDIYSFGLIILQLLTGRTPVGLAVLVRNAV--SCGKLSSIL 509
>TC227461 similar to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (59%)
Length = 1204
Score = 147 bits (370), Expect = 2e-35
Identities = 92/244 (37%), Positives = 135/244 (54%), Gaps = 11/244 (4%)
Frame = +2
Query: 433 QVEVLSKLKHPNLITLIG--VNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
+V L L HP+L+ LIG + + + L+YE++P GSLE+HL R + PL W R++
Sbjct: 8 EVNYLGDLVHPHLVKLIGYCIEDDQRLLVYEFMPRGSLENHLFRR----SLPLPWSIRMK 175
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
IA L FLH +++ D K SNILLDA KLSDFG+ + D +
Sbjct: 176 IALGAAKGLAFLHEEAERPVIYRDFKTSNILLDAEYNAKLSDFGLAK------DGPEGDK 337
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL------GIKN 604
T +++ GT+ Y PE++ TG LTS+SDVYSFG+VLL ++TG+ ++ G N
Sbjct: 338 T---HVSTRVMGTYGYAAPEYVMTGHLTSRSDVYSFGVVLLEMLTGRRSMDKNRPNGEHN 508
Query: 605 EVLYALNNAGGNVK--SVLDP-LAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRV 661
V +A + G + ++DP L G + I A+K H A C + K+RP L SE
Sbjct: 509 LVEWARPHLGERRRFYKLIDPRLEGHFSIKGAQKAAHLAAHCLSRDPKARP-LMSEVVEA 685
Query: 662 LEPM 665
L+P+
Sbjct: 686 LKPL 697
>TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(80%)
Length = 1175
Score = 144 bits (362), Expect = 2e-34
Identities = 91/234 (38%), Positives = 133/234 (55%), Gaps = 12/234 (5%)
Frame = +2
Query: 433 QVEVLSKLKHPNLITLIGV--NQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
++ L +L+HPNL+ LIG E + L+YE++P GS+E+HL R G+ P +W R++
Sbjct: 503 EINYLGQLQHPNLVKLIGYCFEDEHRLLVYEFMPKGSMENHLFRRGSY-FQPFSWSLRMK 679
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
IA L FLHS + H +++ D K SNILLD N KLSDFG+ R D + +
Sbjct: 680 IALGAAKGLAFLHSTE-HKVIYRDFKTSNILLDTNYNAKLSDFGLAR------DGPTGDK 838
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL------GIKN 604
+ +++ GT Y PE+L TG LT+KSDVYSFG+VLL +I+G+ A+ G N
Sbjct: 839 SH---VSTRVMGTRGYAAPEYLATGHLTTKSDVYSFGVVLLEMISGRRAIDKNQPTGEHN 1009
Query: 605 EVLYA---LNNAGGNVKSVLDP-LAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
V +A L+N V V+DP L G + A+ A++C + K RP +
Sbjct: 1010LVEWAKPYLSNK-RRVFRVMDPRLEGQYSQNRAQAAAALAMQCFSVEPKCRPNM 1168
>TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(84%)
Length = 1629
Score = 135 bits (341), Expect = 5e-32
Identities = 87/233 (37%), Positives = 130/233 (55%), Gaps = 11/233 (4%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIG--VNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
+V L + H +L+ LIG + E + L+YE++P GS E+HL R G+ PL+W R++
Sbjct: 442 EVNYLGQHFHSHLVRLIGFCLEDEHRLLVYEFMPRGSWENHLFRRGSY-FQPLSWSLRLK 618
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
+A + L FLHS + +++ D K SN+LLD+ KLSDFG+ + D + +
Sbjct: 619 VALDAAKGLAFLHSAEA-KVIYRDFKTSNVLLDSKYNAKLSDFGLAK------DGPTGDK 777
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL------GIKN 604
+ +++ GT+ Y PE+L TG LT+KSDVYSFG+VLL +++GK A+ G N
Sbjct: 778 SH---VSTRVMGTYGYAAPEYLATGHLTAKSDVYSFGVVLLEMLSGKRAVDKNRPSGQHN 948
Query: 605 EVLYALNNAGGNVK--SVLDP-LAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
V +A K VLD L G + +A KL ALRC + K RP +
Sbjct: 949 LVEWAKPFMANKRKIFRVLDTRLQGQYSTDDAYKLATLALRCLSIESKFRPNM 1107
>TC218979 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associated
receptor-like protein kinase, partial (34%)
Length = 1343
Score = 135 bits (340), Expect = 6e-32
Identities = 86/249 (34%), Positives = 134/249 (53%), Gaps = 12/249 (4%)
Frame = +3
Query: 416 APTVLK--DPQNFNRSLSYQVEV--LSKLKHPNLITLIGVNQE--SKTLIYEYLPNGSLE 469
+P +K P + N +Q EV L K+ H NL +LIG E +K LIYEY+ NG+L+
Sbjct: 255 SPVAVKVLSPSSVNGFQQFQAEVKLLVKVHHKNLTSLIGYCNEGTNKALIYEYMANGNLQ 434
Query: 470 DHLSRNGNNNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTK 529
+HLS ++ + L+W+ R+RIA + L +L + I+H D+K +NILL+ + K
Sbjct: 435 EHLSGK-HSKSTFLSWEDRLRIAVDAALGLEYLQNGCKPPIIHRDVKSTNILLNEHFQAK 611
Query: 530 LSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIV 589
LSDFG+ + + +S +++ GT Y+DP + LT KSDVYSFG+V
Sbjct: 612 LSDFGLSKAIPTDGESH---------VSTVLAGTPGYLDPHCHISSRLTQKSDVYSFGVV 764
Query: 590 LLRLITGKPALGIKNEVLYALNNA-----GGNVKSVLDP-LAGDWPIVEAEKLVHFALRC 643
LL +IT +P + E + G++++++D L GD+ I A K + A+ C
Sbjct: 765 LLEIITNQPVMERNQEKGHIRERVRSLIEKGDIRAIVDSRLEGDYDINSAWKALEIAMAC 944
Query: 644 CDMNKKSRP 652
N RP
Sbjct: 945 VSQNPNERP 971
>TC207707 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase-like
protein, partial (34%)
Length = 1037
Score = 134 bits (336), Expect = 2e-31
Identities = 71/169 (42%), Positives = 103/169 (60%), Gaps = 2/169 (1%)
Frame = +1
Query: 434 VEVLSKLKHPNLITLIGVNQESKT--LIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRI 491
VE+LSKL+H +L++LIG E L+YEY+ NG L HL + PPL+W+ R+ I
Sbjct: 1 VEMLSKLRHCHLVSLIGYCDERSEMILVYEYMANGPLRSHLY---GTDLPPLSWKQRLEI 171
Query: 492 ATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNST 551
L +LH+ SI+H D+K +NILLD N V K++DFG+ S + S
Sbjct: 172 CIGAARGLHYLHTGAAQSIIHRDVKTTNILLDENFVAKVADFGL---------SKTGPSL 324
Query: 552 TQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL 600
Q +++ KG+F Y+DPE+ +LT KSDVYSFG+VL+ ++ +PAL
Sbjct: 325 DQTHVSTAVKGSFGYLDPEYFKRQQLTEKSDVYSFGVVLMEVLCTRPAL 471
>TC226831 similar to PIR|T52285|T52285 serine/threonine-specific protein
kinase APK2a [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (82%)
Length = 1693
Score = 133 bits (335), Expect = 2e-31
Identities = 91/257 (35%), Positives = 141/257 (54%), Gaps = 11/257 (4%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIG--VNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
+V+ L +L H NL+ LIG + E++ L+YE++ GSLE+HL R G PL+W R++
Sbjct: 637 EVDYLGQLHHQNLVKLIGYCADGENRLLVYEFMSKGSLENHLFRRGPQ---PLSWSVRMK 807
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
+A L FLH+ K +++ D K SNILLDA KLSDFG+ + + +
Sbjct: 808 VAIGAARGLSFLHNAKSQ-VIYRDFKASNILLDAEFNAKLSDFGLAK---------AGPT 957
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL-----GI-KN 604
+ +++ GT Y PE++ TG LT+KSDVYSFG+VLL L++G+ A+ G+ +N
Sbjct: 958 GDRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDRSKAGVEQN 1137
Query: 605 EVLYALNNAGGNVK--SVLD-PLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRV 661
V +A G + ++D L G +P A AL+C + K RP + +E +
Sbjct: 1138LVEWAKPYLGDKRRLFRIMDTKLGGQYPQKGAYMAATLALKCLNREAKGRPPI-TEVLQT 1314
Query: 662 LEPMKVSCSGTNNFGLK 678
LE + S + N L+
Sbjct: 1315LEQIAASKTAGRNSQLE 1365
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 133 bits (334), Expect = 3e-31
Identities = 91/246 (36%), Positives = 139/246 (55%), Gaps = 12/246 (4%)
Frame = +2
Query: 433 QVEVLSKLKHPNLITLIGV--NQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
++ L +L+HP+L+ LIG E + L+YEY+P GSLE+ L R + P W TR++
Sbjct: 224 EIIFLGQLRHPHLVKLIGYCYEDEHRLLMYEYMPRGSLENQLFRRYSAAMP---WSTRMK 394
Query: 491 IATELCSALIFLH-SNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNN 549
IA L FLH ++KP +++ D K SNILLD++ KLSDFG+ + D
Sbjct: 395 IALGAAKGLAFLHEADKP--VIYRDFKASNILLDSDFTAKLSDFGLAK------DGPEGE 550
Query: 550 STTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGI------K 603
T +T+ GT Y PE++ TG LT+KSDVYS+G+VLL L+TG+ + K
Sbjct: 551 DTH---VTTRIMGTQGYAAPEYIMTGHLTTKSDVYSYGVVLLELLTGRRVVDKSRSNEGK 721
Query: 604 NEVLYA--LNNAGGNVKSVLD-PLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWR 660
+ V +A L V +++D L G +P+ A K+ A +C + +RP + S+ +
Sbjct: 722 SLVEWARPLLRDQKKVYNIIDRRLEGQFPMKGAMKVAMLAFKCLSHHPNARPTM-SDVIK 898
Query: 661 VLEPMK 666
VLEP++
Sbjct: 899 VLEPLQ 916
>TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(78%)
Length = 1607
Score = 132 bits (333), Expect = 4e-31
Identities = 83/233 (35%), Positives = 131/233 (55%), Gaps = 11/233 (4%)
Frame = +3
Query: 433 QVEVLSKLKHPNLITLIGVNQESKT--LIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
+V L +L HP+L+ LIG E K L+YE++P GSLE+HL G+ PL+W R++
Sbjct: 507 EVNYLGQLSHPHLVKLIGYCFEDKDRLLVYEFMPRGSLENHLFMRGSY-FQPLSWGLRLK 683
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
+A L FLHS + +++ D K SN+LLD+N KL+D G+ + + S ++
Sbjct: 684 VALGAAKGLAFLHSAET-KVIYRDFKTSNVLLDSNYNAKLADLGLAKDGPTREKSHASTR 860
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL------GIKN 604
GT+ Y PE+L TG L++KSDV+SFG+VLL +++G+ A+ G N
Sbjct: 861 VM---------GTYGYAAPEYLATGNLSAKSDVFSFGVVLLEMLSGRRAVDKNRPSGQHN 1013
Query: 605 EVLYA---LNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
V +A L+N ++ + + L G + + EA K+ +LRC + K RP +
Sbjct: 1014LVEWAKPYLSNKRKLLRVLDNRLEGQYELDEACKVATLSLRCLAIESKLRPTM 1172
>TC219331 similar to UP|Q9FM85 (Q9FM85) Protein kinase-like protein
(At5g56460), partial (61%)
Length = 1183
Score = 132 bits (331), Expect = 7e-31
Identities = 88/232 (37%), Positives = 127/232 (53%), Gaps = 12/232 (5%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIGVNQES--KTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
QVE +L HPNL+ +IG E + LIYEY+ G L+++L + PPL+W R++
Sbjct: 124 QVEFWGQLSHPNLVKVIGYCCEDNHRVLIYEYMSRGGLDNYLFKYAPA-IPPLSWSMRMK 300
Query: 491 IATELCSALIFLH-SNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNN 549
IA L FLH + KP +++ D K SNILLD +KLSDFG+ + + S
Sbjct: 301 IAFGAAKGLAFLHEAEKP--VIYRDFKTSNILLDQEYNSKLSDFGLAKDGPVGDKSH--- 465
Query: 550 STTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALG------IK 603
+++ GT+ Y PE++ TG LT +SDVYSFG+VLL L+TG+ +L +
Sbjct: 466 ------VSTRVMGTYGYAAPEYIMTGHLTPRSDVYSFGVVLLELLTGRKSLDKLRPAREQ 627
Query: 604 NEVLYALNNAGGNVK--SVLDP-LAGDWPIVEAEKLVHFALRCCDMNKKSRP 652
N +AL K +++DP L GD+PI K A C + N K+RP
Sbjct: 628 NLAEWALPLLKEKKKFLNIIDPRLDGDYPIKAVHKAAMLAYHCLNRNPKARP 783
>TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase-like
protein, partial (38%)
Length = 1357
Score = 132 bits (331), Expect = 7e-31
Identities = 68/170 (40%), Positives = 104/170 (61%), Gaps = 2/170 (1%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIGVNQESKT--LIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
++E+LSKL+H +L++LIG E L+YEY+ NG L HL + PPL+W+ R+
Sbjct: 100 EIEMLSKLRHRHLVSLIGYCDERSEMILVYEYMANGPLRSHLY---GTDLPPLSWKQRLE 270
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
I L +LH+ SI+H D+K +NIL+D N V K++DFG+ S + +
Sbjct: 271 ICIGAARGLHYLHTGASQSIIHRDVKTTNILVDDNFVAKVADFGL---------SKTGPA 423
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL 600
Q +++ KG+F Y+DPE+ +LT KSDVYSFG+VL+ ++ +PAL
Sbjct: 424 LDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLMEVLCTRPAL 573
>TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like protein,
partial (66%)
Length = 1737
Score = 131 bits (330), Expect = 9e-31
Identities = 86/261 (32%), Positives = 136/261 (51%), Gaps = 13/261 (4%)
Frame = +3
Query: 433 QVEVLSKLKHPNLITLIGVNQESKT--LIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
++E+LSKL+H +L++LIG +E+ L+Y+Y+ G+L +HL + PP W+ R+
Sbjct: 570 EIEMLSKLRHRHLVSLIGYCEENTEMILVYDYMAYGTLREHLYKT---QKPPRPWKQRLE 740
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
I L +LH+ H+I+H D+K +NILLD V K+SDFG+ S + +
Sbjct: 741 ICIGAARGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGL---------SKTGPT 893
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALG---IKNEVL 607
+++ KG+F Y+DPE+ +LT KSDVYSFG+VL ++ +PAL K +V
Sbjct: 894 LDNTHVSTVVKGSFGYLDPEYFRRQQLTDKSDVYSFGVVLFEVLCARPALNPTLAKEQVS 1073
Query: 608 YALNNA----GGNVKSVLDP-LAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRV- 661
A A G + S++DP L G +K A++C RP + W +
Sbjct: 1074LAEWAAHCYQKGILDSIIDPYLKGKIAPECFKKFAETAMKCVADQGIDRPSMGDVLWNLE 1253
Query: 662 --LEPMKVSCSGTNNFGLKSC 680
L+ + + N FG C
Sbjct: 1254FALQLQESAEESGNGFGDIHC 1316
>TC227460 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (44%)
Length = 581
Score = 131 bits (330), Expect = 9e-31
Identities = 70/169 (41%), Positives = 101/169 (59%), Gaps = 2/169 (1%)
Frame = +3
Query: 434 VEVLSKLKHPNLITLIG--VNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRI 491
V L L HPNL+ L+G + ++ + L+YE++P GSLE+HL R + PL W R++I
Sbjct: 3 VNFLGDLVHPNLVKLVGYCIEEDQRLLVYEFMPRGSLENHLFRR----SIPLPWSIRMKI 170
Query: 492 ATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNST 551
A L FLH +++ D K SNILLDA KLSDFG+ + D + T
Sbjct: 171 ALGAAKGLAFLHEEAERPVIYRDFKTSNILLDAEYNAKLSDFGLAK------DGPEGDKT 332
Query: 552 TQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL 600
+++ GT+ Y PE++ TG LTSKSDVYSFG+VLL ++TG+ ++
Sbjct: 333 H---VSTRVMGTYGYAAPEYVMTGHLTSKSDVYSFGVVLLEMLTGRRSM 470
>TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete
Length = 3053
Score = 131 bits (330), Expect = 9e-31
Identities = 67/170 (39%), Positives = 104/170 (60%), Gaps = 2/170 (1%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIG--VNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
++ VLSK++H +L+ L+G +N + L+YEY+P G+L HL PLTW+ R+
Sbjct: 1768 EIAVLSKVRHRHLVALLGYCINGIERLLVYEYMPQGTLTQHLFEWQEQGYVPLTWKQRVV 1947
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
IA ++ + +LHS S +H DLKPSNILL ++ K++DFG+ + N
Sbjct: 1948 IALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVK----------NAP 2097
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL 600
++ + + GTF Y+ PE+ TG +T+K D+Y+FGIVL+ LITG+ AL
Sbjct: 2098 DGKYSVETRLAGTFGYLAPEYAATGRVTTKVDIYAFGIVLMELITGRKAL 2247
>TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial (5%)
Length = 1279
Score = 131 bits (330), Expect = 9e-31
Identities = 81/245 (33%), Positives = 129/245 (52%), Gaps = 9/245 (3%)
Frame = +1
Query: 419 VLKDPQNFNRSLSYQVEVLSKLKHPNLITLIGV--NQESKTLIYEYLPNGSLEDHLSRNG 476
++K + F+R ++E+L +KH L+ L G + SK LIY+YLP GSL++ L
Sbjct: 13 IVKLNEGFDRFFERELEILGSIKHRYLVNLRGYCNSPTSKLLIYDYLPGGSLDEAL---- 180
Query: 477 NNNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGIC 536
+ A L W +R+ I L +LH + I+H D+K SNILLD NL ++SDFG+
Sbjct: 181 HERADQLDWDSRLNIIMGAAKGLAYLHHDCSPRIIHRDIKSSNILLDGNLEARVSDFGLA 360
Query: 537 RVLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITG 596
++L D S+ IT+ GTF Y+ PE++ +G T KSDVYSFG++ L +++G
Sbjct: 361 KLL---EDEESH-------ITTIVAGTFGYLAPEYMQSGRATEKSDVYSFGVLTLEVLSG 510
Query: 597 K----PALGIKNEVLYALNN---AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKK 649
K A K + N + ++DPL + + L+ A++C + +
Sbjct: 511 KRPTDAAFIEKGPNIVGWLNFLITENRPREIVDPLCEGVQMESLDALLSVAIQCVSSSPE 690
Query: 650 SRPEL 654
RP +
Sbjct: 691 DRPTM 705
>TC229706 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (50%)
Length = 1162
Score = 130 bits (328), Expect = 1e-30
Identities = 85/221 (38%), Positives = 119/221 (53%), Gaps = 9/221 (4%)
Frame = +2
Query: 454 ESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHG 513
+ + L+YE++P GSLE+HL R PL W R++IA L FLH I++
Sbjct: 8 DQRLLVYEFMPRGSLENHLFRR----PLPLPWSIRMKIALGAAKGLAFLHEEAQRPIIYR 175
Query: 514 DLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLG 573
D K SNILLDA KLSDFG+ + D T +++ GT+ Y PE++
Sbjct: 176 DFKTSNILLDAEYNAKLSDFGLAK------DGPEGEKTH---VSTRVMGTYGYAAPEYVM 328
Query: 574 TGELTSKSDVYSFGIVLLRLITGKPAL------GIKNEVLYALNNAGGN--VKSVLDP-L 624
TG LTSKSDVYSFG+VLL ++TG+ ++ G N V +A G ++DP L
Sbjct: 329 TGHLTSKSDVYSFGVVLLEMLTGRRSIDKKRPNGEHNLVEWARPVLGDRRMFYRIIDPRL 508
Query: 625 AGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPM 665
G + + A+K A +C + KSRP L SE R L+P+
Sbjct: 509 EGHFSVKGAQKAALLAAQCLSRDPKSRP-LMSEVVRALKPL 628
>TC218803 similar to UP|Q84P72 (Q84P72) Receptor-like protein kinase-like
protein (Fragment), partial (35%)
Length = 1266
Score = 130 bits (327), Expect = 2e-30
Identities = 66/170 (38%), Positives = 111/170 (64%), Gaps = 2/170 (1%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIGVNQES--KTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
++ VLSK++H +L++L+G + E + L+YEY+P G+L HL +++ PL+W+ R+
Sbjct: 100 EIAVLSKVRHRHLVSLLGYSTEGNERILVYEYMPQGALSKHLFHWKSHDLEPLSWKRRLN 279
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
IA ++ + +LH+ S +H DLKPSNILL + K+SDFG+ + L+ + + +S
Sbjct: 280 IALDVARGMEYLHTLAHQSFIHRDLKPSNILLADDFKAKVSDFGLVK-LAPEGEKAS--- 447
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL 600
+ + GTF Y+ PE+ TG++T+K+DV+SFG+VL+ L+TG AL
Sbjct: 448 -----VVTRLAGTFGYLAPEYAVTGKITTKADVFSFGVVLMELLTGLMAL 582
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 129 bits (325), Expect = 3e-30
Identities = 83/233 (35%), Positives = 123/233 (52%), Gaps = 13/233 (5%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIGVNQES--KTLIYEYLPNGSLEDHLSRNGNNNAPP--LTWQTR 488
+VE+LS+L P L+ L+G +S K L+YE++ NG L++HL N+ P L W+TR
Sbjct: 130 EVELLSRLHSPYLLALLGYCSDSNHKLLVYEFMANGGLQEHLYPVSNSIITPVKLDWETR 309
Query: 489 IRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSN 548
+RIA E L +LH + ++H D K SNILLD K+SDFG+ ++ D +
Sbjct: 310 LRIALEAAKGLEYLHEHVSPPVIHRDFKSSNILLDKKFHAKVSDFGLAKL---GPDRAGG 480
Query: 549 NSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGK--------PAL 600
+ +T+ GT Y+ PE+ TG LT+KSDVYS+G+VLL L+TG+ P
Sbjct: 481 HVSTR------VLGTQGYVAPEYALTGHLTTKSDVYSYGVVLLELLTGRVPVDMKRPPGE 642
Query: 601 GIKNEVLYALNNAGGNVKSVLDP-LAGDWPIVEAEKLVHFALRCCDMNKKSRP 652
G+ L V ++DP L G + + E ++ A C RP
Sbjct: 643 GVLVSWALPLLTDREKVVKIMDPSLEGQYSMKEVVQVAAIAAMCVQPEADYRP 801
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.331 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,620,976
Number of Sequences: 63676
Number of extensions: 438746
Number of successful extensions: 4777
Number of sequences better than 10.0: 861
Number of HSP's better than 10.0 without gapping: 4066
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4123
length of query: 688
length of database: 12,639,632
effective HSP length: 104
effective length of query: 584
effective length of database: 6,017,328
effective search space: 3514119552
effective search space used: 3514119552
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146341.5


