

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.5 + phase: 0
(220 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
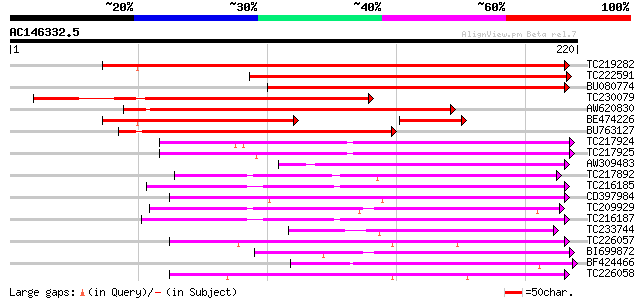
Score E
Sequences producing significant alignments: (bits) Value
TC219282 weakly similar to UP|Q9FFZ7 (Q9FFZ7) Gb|AAF00668.1, par... 214 3e-56
TC222591 weakly similar to GB|AAO23628.1|27765014|BT003063 At2g2... 168 2e-42
BU080774 135 1e-32
TC230079 similar to UP|Q9FFZ7 (Q9FFZ7) Gb|AAF00668.1, partial (45%) 128 2e-30
AW620830 108 1e-24
BE474226 89 2e-23
BU763127 94 5e-20
TC217924 76 1e-14
TC217925 75 2e-14
AW309483 74 4e-14
TC217892 similar to UP|Q6I7V6 (Q6I7V6) ATPase subunit 6, partial... 70 8e-13
TC216185 67 5e-12
CD397984 67 6e-12
TC209929 weakly similar to GB|AAM70533.1|21700819|AY124824 At2g3... 65 3e-11
TC216187 64 7e-11
TC233744 58 4e-09
TC226057 weakly similar to GB|AAG40073.2|13265596|AF324722 AT4g1... 55 3e-08
BI699872 54 6e-08
BF424466 54 6e-08
TC226058 similar to GB|AAG40073.2|13265596|AF324722 AT4g15610 {A... 54 7e-08
>TC219282 weakly similar to UP|Q9FFZ7 (Q9FFZ7) Gb|AAF00668.1, partial (82%)
Length = 751
Score = 214 bits (544), Expect = 3e-56
Identities = 108/184 (58%), Positives = 135/184 (72%), Gaps = 3/184 (1%)
Frame = +1
Query: 37 PHAATVVTTKAT---PLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQ 93
P ++ V KA P + GG KKG+AI+DFILRLGAI AALGAA MGT++Q LPFFTQ
Sbjct: 73 PESSNVAKGKAVLAAPPRPGGWKKGVAIMDFILRLGAIAAALGAAATMGTSDQTLPFFTQ 252
Query: 94 FLQFHAQWDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMA 153
F QF A +D F F+FFV+ G+L+LSLPFS V I+RP AAGPR L+I+D V +
Sbjct: 253 FFQFEASYDSFTSFQFFVITMALVGGYLVLSLPFSFVAIIRPHAAGPRLFLIILDTVFLT 432
Query: 154 LVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVS 213
L A+ +SAAA+VYLAHNG+QD+NW AIC QF DFC +S AVV+SFVA V L LVV+S
Sbjct: 433 LATASGASAAAIVYLAHNGNQDSNWLAICNQFGDFCAQTSSAVVSSFVAVVVLVLLVVMS 612
Query: 214 SVAL 217
+++L
Sbjct: 613 ALSL 624
>TC222591 weakly similar to GB|AAO23628.1|27765014|BT003063 At2g27370
{Arabidopsis thaliana;} , partial (45%)
Length = 459
Score = 168 bits (425), Expect = 2e-42
Identities = 80/125 (64%), Positives = 97/125 (77%)
Frame = +2
Query: 94 FLQFHAQWDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMA 153
FLQFHAQW DFP+F+FFV ANG +G+ ILSLPFS VCIV+P A PR LL+ D V+M
Sbjct: 2 FLQFHAQWSDFPVFQFFVFANGVISGYAILSLPFSYVCIVQPHAVRPRLLLMTFDTVMMG 181
Query: 154 LVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVS 213
L+ AA+ AAA+VY+ HNGSQDANW A CQ FT+FCQ +S AVV SFVA+ F CLV +S
Sbjct: 182 LISVAAAGAAAIVYVGHNGSQDANWMAFCQGFTNFCQAASEAVVLSFVAAAFFLCLVPLS 361
Query: 214 SVALK 218
++ALK
Sbjct: 362 ALALK 376
>BU080774
Length = 436
Score = 135 bits (341), Expect = 1e-32
Identities = 64/117 (54%), Positives = 83/117 (70%)
Frame = +1
Query: 101 WDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAAS 160
+D F F+FFV+ G+L+LSLPFSIV IVRP A GPR L+I+D + L + +
Sbjct: 28 YDSFTTFQFFVITMSLVGGYLVLSLPFSIVAIVRPHAVGPRLFLIILDTAFLTLATSGGA 207
Query: 161 SAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
SAAA+VYLAHNG QD NW AIC QF DFC +S AVV+SFVA V L L+++S++A+
Sbjct: 208 SAAAIVYLAHNGDQDTNWLAICNQFGDFCAQTSAAVVSSFVAVVVLVLLIIMSALAI 378
>TC230079 similar to UP|Q9FFZ7 (Q9FFZ7) Gb|AAF00668.1, partial (45%)
Length = 450
Score = 128 bits (322), Expect = 2e-30
Identities = 72/132 (54%), Positives = 83/132 (62%)
Frame = +2
Query: 10 SSTAPILESKRTRSNGKGKSIDGDHSPPHAATVVTTKATPLQKGGMKKGIAILDFILRLG 69
S+T I ES + GKG VV A P GG KKG+AI+DFILRLG
Sbjct: 95 STTIEIPESSTKVAKGKG-------------VVVVAPARP---GGWKKGVAIMDFILRLG 226
Query: 70 AIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFLILSLPFSI 129
AI AALGAA MGT++Q LPFFTQF QF A +D F F+FFV+ G+L+LSLPFSI
Sbjct: 227 AIAAALGAAATMGTSDQTLPFFTQFFQFEASYDSFTTFQFFVITMALVGGYLVLSLPFSI 406
Query: 130 VCIVRPLAAGPR 141
V IVRP A GPR
Sbjct: 407 VAIVRPHAVGPR 442
>AW620830
Length = 430
Score = 108 bits (271), Expect = 1e-24
Identities = 59/129 (45%), Positives = 82/129 (62%)
Frame = +2
Query: 45 TKATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDF 104
+K P +KG + +G++I+DFILR+ A A LG+A+ MGT Q LPF TQF++F A + D
Sbjct: 44 SKGAPPRKG-LIRGLSIMDFILRIVAAIATLGSALGMGTTRQTLPFSTQFVKFRAVFSDL 220
Query: 105 PMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAA 164
P F FFV +N G+L+LSL S IVR A + L V +D V+ L+ AS+A A
Sbjct: 221 PTFVFFVTSNSIVCGYLVLSLVLSFFHIVRSAAVKSKVLQVFLDTVMYGLLTTGASAATA 400
Query: 165 VVYLAHNGS 173
+VY AH G+
Sbjct: 401 IVYEAHYGN 427
>BE474226
Length = 416
Score = 88.6 bits (218), Expect(2) = 2e-23
Identities = 45/80 (56%), Positives = 56/80 (69%), Gaps = 4/80 (5%)
Frame = +3
Query: 37 PHAATVVTTKAT----PLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFT 92
P ++ V KA P + GG KKG+AI+DFILRLGAI AALGAA MGT++Q LPFFT
Sbjct: 105 PESSKVAKGKAVAVVAPARPGGWKKGVAIMDFILRLGAIAAALGAAATMGTSDQTLPFFT 284
Query: 93 QFLQFHAQWDDFPMFKFFVV 112
QF QF A +D F F+ F++
Sbjct: 285 QFFQFEASYDSFTTFQRFLL 344
Score = 37.7 bits (86), Expect(2) = 2e-23
Identities = 16/26 (61%), Positives = 19/26 (72%)
Frame = +2
Query: 152 MALVVAAASSAAAVVYLAHNGSQDAN 177
+ L A +SAAA+VYLAHNG QD N
Sbjct: 338 LTLATAGGASAAAIVYLAHNGDQDTN 415
>BU763127
Length = 426
Score = 94.0 bits (232), Expect = 5e-20
Identities = 49/108 (45%), Positives = 71/108 (65%)
Frame = +3
Query: 43 VTTKATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWD 102
V+ A+P + G+ +G++I+DFILR+ A A LG+A+ M T + LPF TQF++F A++D
Sbjct: 93 VSKDASP--RKGVARGLSIMDFILRIIAAVATLGSALAMSTTNETLPFATQFIKFRAEFD 266
Query: 103 DFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLV 150
D P FFV+AN G+L+LSL S+ I+R A R LLV +D V
Sbjct: 267 DLPSLVFFVMANAVVCGYLVLSLMISVFHILRSTAVKSRILLVALDTV 410
>TC217924
Length = 843
Score = 75.9 bits (185), Expect = 1e-14
Identities = 48/169 (28%), Positives = 84/169 (49%), Gaps = 8/169 (4%)
Frame = +3
Query: 59 IAILDFILRLGAIGAALGAAVIMGTNEQ--ILP------FFTQFLQFHAQWDDFPMFKFF 110
+ + D +LRL A L AA+++ ++Q ++P F + A+W F +F
Sbjct: 183 VGVCDLLLRLLAFTVTLVAAIVIAVDKQTKLVPIQLSDSFPPLNVPLTAKWHQMSAFVYF 362
Query: 111 VVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAH 170
+V N A + +SL ++V R + G L+ ++D ++AL+ + +AAAV L +
Sbjct: 363 LVTNAIACTYAAMSLLLALVN--RGKSKGLWTLIAVLDTFMVALLFSGNGAAAAVGILGY 536
Query: 171 NGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
G+ NWN +C F FC + ++ S + S+ LVV+ V L R
Sbjct: 537 KGNSHVNWNKVCNVFGKFCDQMAASIGVSLIGSLAFLLLVVIPVVRLHR 683
>TC217925
Length = 805
Score = 75.5 bits (184), Expect = 2e-14
Identities = 46/169 (27%), Positives = 81/169 (47%), Gaps = 8/169 (4%)
Frame = +3
Query: 59 IAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQF--------LQFHAQWDDFPMFKFF 110
+ + D +LRL A L AA+++ ++Q Q + A+W +F
Sbjct: 66 VGVSDLLLRLLAFTVTLVAAIVIAVDKQTKVVPIQLSDSLPPLDVPLTAKWHQMSAIVYF 245
Query: 111 VVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAH 170
+V N A + +LSL ++V R + G L+ ++D ++AL+ + +AAAV L +
Sbjct: 246 LVTNAIACTYAVLSLLLALVN--RGKSKGLWTLIAVLDAFMVALLFSGNGAAAAVGVLGY 419
Query: 171 NGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
G+ NWN +C F FC + ++ S + S+ LV++ V L R
Sbjct: 420 KGNSHVNWNKVCNVFGKFCDQMAASIGVSLIGSLAFLLLVIIPGVRLHR 566
>AW309483
Length = 340
Score = 74.3 bits (181), Expect = 4e-14
Identities = 46/113 (40%), Positives = 62/113 (54%)
Frame = -1
Query: 105 PMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAA 164
P+F F G G L S PF V I P AAGPR L+I+ V + L +S +A
Sbjct: 337 PIFGFHKALEG---GHLCASPPFLFVAINCPQAAGPRLFLIILSPVFLTLATGNGASTSA 167
Query: 165 VVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
VYLA + + + NW + F FC +S VV+SFVA V L LVV+S+++L
Sbjct: 166 NVYLATHWNPNLNWVSNSHPFGKFCAQTSSTVVSSFVAVVVLVLLVVMSALSL 8
>TC217892 similar to UP|Q6I7V6 (Q6I7V6) ATPase subunit 6, partial (10%)
Length = 989
Score = 70.1 bits (170), Expect = 8e-13
Identities = 43/152 (28%), Positives = 76/152 (49%), Gaps = 2/152 (1%)
Frame = +1
Query: 65 ILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFLILS 124
ILR+ + + +M TN Q + F ++F A + P FKFFV ANG +L+
Sbjct: 232 ILRIIPTLLSAASIAVMVTNNQTVLIFA--IRFQAHFYYSPSFKFFVAANGVVVALSLLT 405
Query: 125 LPFSIVCIVRPLAAGPR--FLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAIC 182
L ++ + A PR F L + D+V+M L++A ++A A+ Y+ G W IC
Sbjct: 406 L---VLNFLMKRQASPRYHFFLFLHDIVMMVLLIAGCAAATAIGYVGQFGEDHVGWQPIC 576
Query: 183 QQFTDFCQGSSLAVVASFVASVFLACLVVVSS 214
FC + ++++ S+ A + + ++S+
Sbjct: 577 DHVRKFCTTNLVSLLLSYFAFISYFGITLLSA 672
>TC216185
Length = 1098
Score = 67.4 bits (163), Expect = 5e-12
Identities = 46/164 (28%), Positives = 74/164 (45%)
Frame = +1
Query: 54 GMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVA 113
G + + LRL +G + A V+M N Q + + + D F++ V A
Sbjct: 289 GNTSALRTAETFLRLVPVGLCVSALVLMLKNSQQNEYGS------VDYSDLGAFRYLVHA 450
Query: 114 NGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGS 173
NG AG+ + S I + P + ++D VL +++AA + + V+YLA NG
Sbjct: 451 NGICAGYSLFSAV--IAAMPCPSTIPRAWTFFLLDQVLTYIILAAGAVSTEVLYLAENGD 624
Query: 174 QDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
W++ C F FC + +V +FVA L +VSS L
Sbjct: 625 AATTWSSACGSFGRFCHKVTASVAITFVAVFCYVLLSLVSSYKL 756
>CD397984
Length = 643
Score = 67.0 bits (162), Expect = 6e-12
Identities = 44/158 (27%), Positives = 74/158 (45%), Gaps = 3/158 (1%)
Frame = -3
Query: 63 DFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHA--QWDDFPMFKFFVVANGAAAGF 120
D +LR + L A +I+G + + +QF A +W+ FF+V N A +
Sbjct: 626 DLLLRFLGLSLTLVAVIIVGVDNETKVISYAEMQFKATAKWEYVSAIVFFLVINAIACSY 447
Query: 121 LILSLPFSIVCIVRPLAAGPRFL-LVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWN 179
SL +++ L L +DLV+MAL+ +A +A AV +A G+ W
Sbjct: 446 AAASLVITLMARSHGRKNDVTLLGLTALDLVMMALLFSANGAACAVGVIAQKGNSHVQWM 267
Query: 180 AICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
+C F +C+ + A+V S + S LV++S + L
Sbjct: 266 KVCNVFDAYCRHVTAALVLSIIGSTVFLLLVLLSVLKL 153
>TC209929 weakly similar to GB|AAM70533.1|21700819|AY124824
At2g35760/T20F21.5 {Arabidopsis thaliana;} , partial
(74%)
Length = 1070
Score = 64.7 bits (156), Expect = 3e-11
Identities = 45/171 (26%), Positives = 80/171 (46%), Gaps = 10/171 (5%)
Frame = +1
Query: 55 MKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVAN 114
+ + + I + +LR ++G A V++ T+ Q+ FF+ Q A++ D F VVAN
Sbjct: 463 LDRRVRITELVLRCVSLGLGAVAIVLVVTDSQVKEFFS--FQKKAKFTDMKALVFLVVAN 636
Query: 115 GAAAGFLILSLPFSIVCIVR-------PLAAGPRFLLVIVDLVLMALVVAAASSAAAVVY 167
G G+ ++ +V ++R PLA +L+ D V+ + VAA ++A
Sbjct: 637 GLTVGYSLIQGLRCVVSMIRGNVLFNKPLA----WLIFSGDQVMAYVTVAAVAAALQSGV 804
Query: 168 LAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVAS---VFLACLVVVSSV 215
L G + W +C + FC + ++FV S V L+C+ S+
Sbjct: 805 LGRTGQAELQWMKVCNMYGKFCNQMGEGIASAFVVSLSKVVLSCISAFKSL 957
>TC216187
Length = 977
Score = 63.5 bits (153), Expect = 7e-11
Identities = 43/166 (25%), Positives = 74/166 (43%)
Frame = +1
Query: 52 KGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFV 111
+G + + LRL +G + A V+M + Q + + + D F++ V
Sbjct: 184 EGNTTSALRTAETFLRLFPVGLCVSALVLMLKSSQQNEYGS------VDYSDLGAFRYLV 345
Query: 112 VANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHN 171
ANG AG+ + S I + P + ++D VL +++AA + + V+YLA
Sbjct: 346 HANGICAGYSLFSAV--IAAMPCPSTIPRAWTFFLLDQVLTYIILAAGAVSTEVLYLAEK 519
Query: 172 GSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
G W++ C F FC + +V +FVA L ++SS L
Sbjct: 520 GDAATTWSSACGSFGRFCHKVTASVAITFVAVFCYVLLSLISSYKL 657
>TC233744
Length = 488
Score = 57.8 bits (138), Expect = 4e-09
Identities = 32/113 (28%), Positives = 57/113 (50%), Gaps = 8/113 (7%)
Frame = +3
Query: 109 FFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRF--------LLVIVDLVLMALVVAAAS 160
FFV+ N A+ + ++ + I+ GP++ L+ I+D++ MAL
Sbjct: 6 FFVIVNAIASFYNMVVIGVEIL--------GPQYDYKELRLGLIAILDVMTMALAATGDG 161
Query: 161 SAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVS 213
+A + L NG+ A W+ IC +F +C +A++ASFV + L + V+S
Sbjct: 162 AATFMAELGRNGNSHARWDKICDKFEAYCNRGGVALIASFVGLILLLVVTVMS 320
>TC226057 weakly similar to GB|AAG40073.2|13265596|AF324722 AT4g15610
{Arabidopsis thaliana;} , partial (47%)
Length = 758
Score = 54.7 bits (130), Expect = 3e-08
Identities = 39/158 (24%), Positives = 72/158 (44%), Gaps = 3/158 (1%)
Frame = +1
Query: 63 DFILRLGAIGAALGAAVIMGTNEQI-LPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFL 121
D ILR A+L A V++ T Q + Q + + A++ P F +FV A +
Sbjct: 220 DVILRFVLFAASLVAVVVIVTGNQTEVILVPQPVPWPAKFRYTPAFVYFVAALSVTGLYS 399
Query: 122 ILSLPFSIVCIVRPLAAGPRFLLVIV-DLVLMALVVAAASSAAAVVYLAHNG-SQDANWN 179
I++ S+ +P L I+ D +++ ++ +A +A V YL G S WN
Sbjct: 400 IITTLASLFASNKPALKTKLLLYFILWDALILGIIASATGTAGGVAYLGLKGNSHVVGWN 579
Query: 180 AICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
IC + FC+ ++ + S+ L+ +S+ ++
Sbjct: 580 KICHVYDKFCRHVGASIAVALFGSIVTVLLIWLSAYSI 693
>BI699872
Length = 420
Score = 53.9 bits (128), Expect = 6e-08
Identities = 38/131 (29%), Positives = 61/131 (46%), Gaps = 7/131 (5%)
Frame = +2
Query: 96 QFHAQWDDFPMFKFFVVANGAAAGF-------LILSLPFSIVCIVRPLAAGPRFLLVIVD 148
Q A++ D F VVANG AAG+ ILS+ V +PLA + + D
Sbjct: 14 QKEAKFTDMKSLVFLVVANGLAAGYSLIQGLRCILSMIRGRVLFSKPLA----WAIFSGD 181
Query: 149 LVLMALVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLAC 208
V+ + V A ++A +A G + W IC + FC + ++FVAS+ +
Sbjct: 182 QVMAYVTVVALAAAGQSGMIARVGQPELQWMKICNMYGKFCNQVGEGIASAFVASLSMVV 361
Query: 209 LVVVSSVALKR 219
+ +S+ +L R
Sbjct: 362 MSCISAFSLFR 394
>BF424466
Length = 419
Score = 53.9 bits (128), Expect = 6e-08
Identities = 34/112 (30%), Positives = 58/112 (51%), Gaps = 1/112 (0%)
Frame = +1
Query: 110 FVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLA 169
+V AN A + ILSL ++ R L+ ++D V++AL+ + +A AV L
Sbjct: 4 YVGANAIACAYAILSLLLTLANR-RKGKGTMETLITVLDTVMVALLFSGNGAAMAVGLLG 180
Query: 170 HNGSQDANWNAICQQFTDFCQGSSLAVVASFVASV-FLACLVVVSSVALKRT 220
G+ +WN +C +F FC + ++ S + S+ FL L+V + LK+T
Sbjct: 181 LQGNSHVHWNKVCNEFGKFCDQVAASLFISLLGSIAFLLLLIVPPVLRLKQT 336
>TC226058 similar to GB|AAG40073.2|13265596|AF324722 AT4g15610 {Arabidopsis
thaliana;} , partial (47%)
Length = 1014
Score = 53.5 bits (127), Expect = 7e-08
Identities = 39/158 (24%), Positives = 74/158 (46%), Gaps = 3/158 (1%)
Frame = +1
Query: 63 DFILRLGAIGAALGAAVIMGT-NEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFL 121
D ILR A+L A V++ T N+ + Q + + A++ P F +FV A +
Sbjct: 250 DVILRFLLFAASLVAVVVIVTANQTEVIRVPQPVPWPAKFRYSPAFVYFVAALSVTGLYS 429
Query: 122 ILSLPFSIVCIVRPLAAGPRFLLVIV-DLVLMALVVAAASSAAAVVYLAHNGSQDA-NWN 179
I++ S+ +P L I+ D +++ ++ +A +A V YL G++ WN
Sbjct: 430 IITTLASLFASNKPALKTKLLLYFILWDALILGIIASATGTAGGVAYLGLKGNRHVVGWN 609
Query: 180 AICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
IC + FC+ ++ + SV L+ +S+ ++
Sbjct: 610 KICHVYDKFCRHVGASIAVALFGSVVTVLLIWLSAYSI 723
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.136 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,969,388
Number of Sequences: 63676
Number of extensions: 117548
Number of successful extensions: 881
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 870
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 874
length of query: 220
length of database: 12,639,632
effective HSP length: 93
effective length of query: 127
effective length of database: 6,717,764
effective search space: 853156028
effective search space used: 853156028
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146332.5


