

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.3 - phase: 0
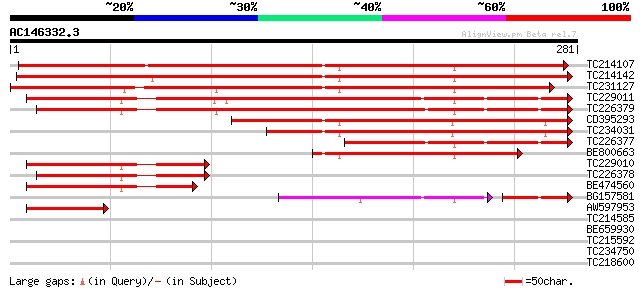
(281 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214107 similar to PIR|T49142|T49142 CCR4-associated factor 1-l... 330 6e-91
TC214142 similar to PIR|T49142|T49142 CCR4-associated factor 1-l... 328 2e-90
TC231127 similar to GB|BAB08323.1|9757805|AB007651 CCR4-associat... 323 7e-89
TC229011 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247... 241 3e-64
TC226379 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247... 229 1e-60
CD395293 similar to GP|9757805|dbj CCR4-associated factor-like p... 193 6e-50
TC234031 similar to GB|BAB08323.1|9757805|AB007651 CCR4-associat... 171 3e-43
TC226377 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247... 105 3e-23
BE800663 similar to GP|9757805|dbj| CCR4-associated factor-like ... 94 7e-20
TC229010 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247... 91 8e-19
TC226378 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247... 84 9e-17
BE474560 80 8e-16
BG157581 62 2e-13
AW597953 57 1e-08
TC214585 weakly similar to UP|PTRB_MORLA (Q59536) Protease II (... 30 1.2
BE659930 28 3.6
TC215592 similar to UP|Q9SSS9 (Q9SSS9) H+-transporting ATP synth... 28 6.1
TC234750 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY... 28 6.1
TC218600 28 6.1
>TC214107 similar to PIR|T49142|T49142 CCR4-associated factor 1-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(84%)
Length = 1003
Score = 330 bits (845), Expect = 6e-91
Identities = 170/275 (61%), Positives = 205/275 (73%), Gaps = 2/275 (0%)
Frame = +2
Query: 5 KEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHL 64
++ + + PI IR+VWA NLE EF+LIR+ + +P++SMDTEFPGV+ P+ DP P++
Sbjct: 38 EDPNPNPNPIVIREVWASNLESEFELIRELIDRYPFISMDTEFPGVIFRPHVDPTKPFNH 217
Query: 65 RHMDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNFKDFDVDRDLQN 124
R+ PS+ Y LK+NVD LNLIQ+GLTLTDA GNLP IWEFNF+DFDV RD
Sbjct: 218 RNR-PSDHYRLLKSNVDALNLIQVGLTLTDAAGNLPDLAGNRSIWEFNFRDFDVARDAYA 394
Query: 125 PDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWVTFHSSYDFGYLVKI 183
PDSI+LLRRQGIDF RN GVDS FA+L + SGLV N VSWVTFHS+YDFGYLVKI
Sbjct: 395 PDSIDLLRRQGIDFARNTADGVDSTCFAELM-MSSGLVCNDAVSWVTFHSAYDFGYLVKI 571
Query: 184 LTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVGNSHQ 242
LT+ LP+RLEEFL + FG NVYD+K+M++FC+ LYGGL+RVA L V RAVG HQ
Sbjct: 572 LTRRNLPTRLEEFLKTVKVFFGDNVYDVKHMMRFCDTLYGGLDRVARTLNVDRAVGKCHQ 751
Query: 243 AASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGL 277
A SDSLLTW AF+K+ DIYFV HAGVLFGL
Sbjct: 752 AGSDSLLTWHAFQKIVDIYFVKEEHRKHAGVLFGL 856
>TC214142 similar to PIR|T49142|T49142 CCR4-associated factor 1-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(85%)
Length = 1439
Score = 328 bits (840), Expect = 2e-90
Identities = 172/281 (61%), Positives = 206/281 (73%), Gaps = 5/281 (1%)
Frame = +1
Query: 4 VKEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYH 63
V E+ + P+ IRQVWA NLE EF LIR+ + +P++SMDTEFPGVV P+ DP PY+
Sbjct: 541 VFSENPNPDPVVIRQVWASNLESEFQLIRELIDHYPFISMDTEFPGVVFRPHLDPTKPYN 720
Query: 64 LRHMDP---SEQYSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNFKDFDVDR 120
R+ + S+ Y LK+NVD LNLIQ+GLTLTDA GNLP IWEFNF+DFDV R
Sbjct: 721 HRNNNRNRHSDHYRLLKSNVDALNLIQVGLTLTDAAGNLPDLAGNRSIWEFNFRDFDVAR 900
Query: 121 DLQNPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWVTFHSSYDFGY 179
D DSI+LLRRQGIDF RN GV+S FA+L + SGLV N VSWVTFHS+YDFGY
Sbjct: 901 DAYALDSIDLLRRQGIDFARNATDGVESTRFAELM-MSSGLVCNDSVSWVTFHSAYDFGY 1077
Query: 180 LVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVG 238
LVKILT+ LP+RLEEFL + FG NVYD+K+M++FC+ LYGGL+RVA L V RAVG
Sbjct: 1078LVKILTRRNLPTRLEEFLKTVKVFFGDNVYDVKHMMRFCDTLYGGLDRVARSLNVERAVG 1257
Query: 239 NSHQAASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
HQA SDSLLTW AF+K+ DIYFV + HAGVL+GLEV
Sbjct: 1258KCHQAGSDSLLTWHAFQKIVDIYFVKDEHRKHAGVLYGLEV 1380
>TC231127 similar to GB|BAB08323.1|9757805|AB007651 CCR4-associated
factor-like protein {Arabidopsis thaliana;} , partial
(82%)
Length = 886
Score = 323 bits (827), Expect = 7e-89
Identities = 170/274 (62%), Positives = 205/274 (74%), Gaps = 4/274 (1%)
Frame = +3
Query: 1 MTKVKEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPN-FDPN 59
M ++E +K I IR+VWA NLE EF LIR + +P++SMDTEFPGVV P+ DP
Sbjct: 78 MGLLEENPNHSKTILIREVWASNLESEFQLIRQVIDDYPFISMDTEFPGVVFRPHTVDPT 257
Query: 60 IPYHLRHMDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLPG-DVAYSYIWEFNFKDFDV 118
PY + PS Y FLK+NVD LNLIQ+GLTL+D+NGNLP A +IWEFNF+DFDV
Sbjct: 258 KPY----LPPSVHYRFLKSNVDALNLIQIGLTLSDSNGNLPHLGTANRFIWEFNFRDFDV 425
Query: 119 DRDLQNPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWVTFHSSYDF 177
+RD PDSI+LLRRQGIDF+RN GVDS FA+L + SGLV N VSWVTFHS+YDF
Sbjct: 426 ERDAHAPDSIDLLRRQGIDFRRNAAEGVDSYLFAELV-MSSGLVCNDSVSWVTFHSAYDF 602
Query: 178 GYLVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRA 236
GYLVKILT+ LPS LEEFL++L FG NVYD+K+M++FC+ LYGGL+R+A LKV RA
Sbjct: 603 GYLVKILTRRSLPSGLEEFLNMLRAFFGNNVYDIKHMMRFCDTLYGGLDRLARTLKVDRA 782
Query: 237 VGNSHQAASDSLLTWQAFKKMKDIYFVNNGITMH 270
VG HQA SDSLLTW AF+KM+DIYFV +G H
Sbjct: 783 VGKCHQAGSDSLLTWHAFQKMRDIYFVTDGPQKH 884
>TC229011 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247p), partial
(69%)
Length = 1165
Score = 241 bits (615), Expect = 3e-64
Identities = 138/281 (49%), Positives = 183/281 (65%), Gaps = 10/281 (3%)
Frame = +2
Query: 9 ASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDPNIPYHLRH 66
A + I+IR+VW +NLE EF LIR+ V +PY++MDTEFPG+V+ P NF + YH
Sbjct: 221 AKSDSIQIREVWNDNLEEEFALIREIVDNYPYIAMDTEFPGIVLRPVGNFKNSYDYH--- 391
Query: 67 MDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLP---GDVAYS----YIWEFNFKDFDVD 119
Y LK NVD L LIQLGLT +D +GNLP GD S IW+FNF++F+V+
Sbjct: 392 ------YQTLKDNVDMLKLIQLGLTFSDEHGNLPMCGGDDEESDTCCCIWQFNFREFNVN 553
Query: 120 RDLQNPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYDFGY 179
D+ DSIELLR+ GIDFKRN G+D+ F +L ++N + WVTFHS YDFGY
Sbjct: 554 EDVFANDSIELLRQSGIDFKRNNENGIDAHRFGELLMSSGIVLNDNIHWVTFHSGYDFGY 733
Query: 180 LVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVG 238
L+K+LT LP F +++ F VYD+K+++KFCN L+GGL ++A L++ R VG
Sbjct: 734 LLKLLTCQDLPDTQVGFFNLINMYF-PTVYDIKHLMKFCNSLHGGLNKLAELLELER-VG 907
Query: 239 NSHQAASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
SHQA SDSLLT FKK KD +F + + +AGVL+GL V
Sbjct: 908 ISHQAGSDSLLTSCTFKKSKDNFF-SGSLEKYAGVLYGLGV 1027
>TC226379 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247p), partial
(73%)
Length = 1380
Score = 229 bits (584), Expect = 1e-60
Identities = 131/270 (48%), Positives = 173/270 (63%), Gaps = 4/270 (1%)
Frame = +3
Query: 14 IKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDPNIPYHLRHMDPSE 71
++IR+VW +NLE EF LIR+ V + YV+MDTEFPGVV+ P NF Y+
Sbjct: 255 VQIREVWNDNLEEEFALIREIVDEYNYVAMDTEFPGVVLRPVGNFKNINDYN-------- 410
Query: 72 QYSFLKANVDNLNLIQLGLTLTDANGNLPG-DVAYSYIWEFNFKDFDVDRDLQNPDSIEL 130
Y LK NVD L LIQLGLT +D NGNLP + IW+FNF++F++ D+ DSIEL
Sbjct: 411 -YQTLKDNVDMLKLIQLGLTFSDENGNLPTCGTESTCIWQFNFREFNISEDIFASDSIEL 587
Query: 131 LRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYDFGYLVKILTQNYLP 190
LR+ GIDFK+N G+D F +L ++N V WVTFHS YDFGYL+K+LT LP
Sbjct: 588 LRQCGIDFKKNSENGIDVNRFGELLMSSGIVLNDAVHWVTFHSGYDFGYLLKLLTCRSLP 767
Query: 191 SRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVGNSHQAASDSLL 249
F ++ F VYD+K+++KFCN L+GGL ++A L+V R VG HQA SDSLL
Sbjct: 768 DTQAGFFDLIKMYFPM-VYDIKHLMKFCNSLHGGLNKLAELLEVER-VGVCHQAGSDSLL 941
Query: 250 TWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
T F+K++D +F + +AGVL+GL V
Sbjct: 942 TSCTFRKLRDTFF-SGSTEKYAGVLYGLGV 1028
>CD395293 similar to GP|9757805|dbj CCR4-associated factor-like protein
{Arabidopsis thaliana}, partial (57%)
Length = 646
Score = 193 bits (491), Expect = 6e-50
Identities = 98/172 (56%), Positives = 129/172 (74%), Gaps = 3/172 (1%)
Frame = -1
Query: 111 FNFKDFDVDRDLQNPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWV 169
FNF+DFDV RD DS+ELLRRQGIDF++N +G+DS FA+L + SGLV ++ VSWV
Sbjct: 625 FNFRDFDVVRDAHAHDSVELLRRQGIDFEKNRDFGIDSFRFAELM-MSSGLVCDNAVSWV 449
Query: 170 TFHSSYDFGYLVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFC-NLYGGLERVA 228
TFHS+YDFGYLVK+LT LP L EFL ++ FG V+D+K++++FC NL+GGL RV
Sbjct: 448 TFHSAYDFGYLVKLLTHRALPEELREFLRLVRVFFGDRVFDVKHLMRFCSNLHGGLNRVC 269
Query: 229 TKLKVSRAVGNSHQAASDSLLTWQAFKKMKDIYFVN-NGITMHAGVLFGLEV 279
LKV R +G SHQA SDSLLT AF+ +++IYF +G+ +AGVL+GL+V
Sbjct: 268 QSLKVERVLGKSHQAGSDSLLTLHAFQNIREIYFGKADGLVQYAGVLYGLDV 113
>TC234031 similar to GB|BAB08323.1|9757805|AB007651 CCR4-associated
factor-like protein {Arabidopsis thaliana;} , partial
(51%)
Length = 472
Score = 171 bits (433), Expect = 3e-43
Identities = 86/155 (55%), Positives = 116/155 (74%), Gaps = 3/155 (1%)
Frame = +2
Query: 128 IELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWVTFHSSYDFGYLVKILTQ 186
+ELLRRQGIDF++N +G+DS FA+L + SGLV + VSWVTFHS+YDFGYLVK++T
Sbjct: 5 VELLRRQGIDFEKNRDFGIDSFRFAELM-MSSGLVCDDAVSWVTFHSAYDFGYLVKLMTH 181
Query: 187 NYLPSRLEEFLSILTQIFGQNVYDMKYMIKFC-NLYGGLERVATKLKVSRAVGNSHQAAS 245
LP L EFL ++ FG V+D+K++++FC NL+GGL+RV LKV R +G SHQA S
Sbjct: 182 RTLPEELREFLRLVRVFFGDRVFDVKHLMRFCSNLHGGLDRVCQSLKVERVIGKSHQAGS 361
Query: 246 DSLLTWQAFKKMKDIYFVN-NGITMHAGVLFGLEV 279
DSLLT AF+ +++ YF +G+ +AGVL+GLEV
Sbjct: 362 DSLLTLHAFQNIRENYFDKADGLVQYAGVLYGLEV 466
>TC226377 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247p), partial
(31%)
Length = 760
Score = 105 bits (261), Expect = 3e-23
Identities = 58/114 (50%), Positives = 77/114 (66%), Gaps = 1/114 (0%)
Frame = +1
Query: 167 SWVTFHSSYDFGYLVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLE 225
SWVTFHS YDFGYL+K+LT LP F ++ F VYD+K+++KFCN L+GGL
Sbjct: 4 SWVTFHSGYDFGYLLKLLTCRSLPETQAGFFDLIKMYFPM-VYDIKHLMKFCNSLHGGLN 180
Query: 226 RVATKLKVSRAVGNSHQAASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
++A L+V R VG HQA SDSLLT F+K++D +F + +AGVL+GL V
Sbjct: 181 KLAELLEVER-VGVCHQAGSDSLLTSCTFRKLRDAFF-SGSTEKYAGVLYGLGV 336
>BE800663 similar to GP|9757805|dbj| CCR4-associated factor-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 316
Score = 94.0 bits (232), Expect = 7e-20
Identities = 52/106 (49%), Positives = 69/106 (65%), Gaps = 2/106 (1%)
Frame = +2
Query: 151 FAKLFRLKSGLV-NSGVSWVTFHSSYDFGYLVKILTQNYLPSRLEEFLSILTQIFGQNVY 209
FA+L + SGL+ N+ +SWVTFHS+Y+F YLVKILT LP+RLE+FL + NVY
Sbjct: 2 FAELM-MSSGLICNNSMSWVTFHSTYNFSYLVKILTHRNLPTRLEKFLKTIKVFCRDNVY 178
Query: 210 DMKYMIKFCN-LYGGLERVATKLKVSRAVGNSHQAASDSLLTWQAF 254
++K++I+F N LY L +A L + N H SDSLLT AF
Sbjct: 179 NIKHIIRFYNTLYNKLNHIAQSLNLKHTRKNYHHTKSDSLLTVHAF 316
>TC229010 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247p), partial
(24%)
Length = 429
Score = 90.5 bits (223), Expect = 8e-19
Identities = 48/93 (51%), Positives = 61/93 (64%), Gaps = 2/93 (2%)
Frame = +3
Query: 9 ASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDPNIPYHLRH 66
A + I+IR+VW +NLE EF LIR+ V +PY++MDTEFPG+V+ P NF + YH
Sbjct: 168 AKSDSIQIREVWNDNLEEEFALIREIVDDYPYIAMDTEFPGIVLRPVGNFKNSYDYH--- 338
Query: 67 MDPSEQYSFLKANVDNLNLIQLGLTLTDANGNL 99
Y LK NVD L LIQLGLT +D +GNL
Sbjct: 339 ------YQTLKDNVDMLKLIQLGLTFSDEHGNL 419
>TC226378 weakly similar to UP|Q9VTS4 (Q9VTS4) CG5684-PA (GH06247p), partial
(24%)
Length = 650
Score = 83.6 bits (205), Expect = 9e-17
Identities = 47/88 (53%), Positives = 57/88 (64%), Gaps = 2/88 (2%)
Frame = +1
Query: 14 IKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDPNIPYHLRHMDPSE 71
++IR+VW +NLE EF LIR+ V + YV+MDTEFPGVV+ P NF Y+
Sbjct: 412 VQIREVWNDNLEEEFALIREIVDEYNYVAMDTEFPGVVLRPVGNFKNINDYN-------- 567
Query: 72 QYSFLKANVDNLNLIQLGLTLTDANGNL 99
Y LK NVD L LIQLGLT +D NGNL
Sbjct: 568 -YQTLKDNVDMLKLIQLGLTFSDENGNL 648
>BE474560
Length = 456
Score = 80.5 bits (197), Expect = 8e-16
Identities = 43/87 (49%), Positives = 55/87 (62%), Gaps = 2/87 (2%)
Frame = +3
Query: 9 ASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDPNIPYHLRH 66
A + I+IR+VW +NLE EF LIR+ V +PY++MDTEFPG+V+ P NF + YH
Sbjct: 222 AKSDSIQIREVWNDNLEEEFALIREIVDNYPYIAMDTEFPGIVLRPVGNFKNSYDYH--- 392
Query: 67 MDPSEQYSFLKANVDNLNLIQLGLTLT 93
Y LK NVD L L QLGLT +
Sbjct: 393 ------YQTLKDNVDMLKLXQLGLTFS 455
>BG157581
Length = 474
Score = 62.4 bits (150), Expect(2) = 2e-13
Identities = 39/110 (35%), Positives = 59/110 (53%), Gaps = 4/110 (3%)
Frame = +3
Query: 134 QGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFH---SSYDFGYLVKILTQNYLP 190
+GIDFKRN G+D+ F +L ++N V WV F + + L + P
Sbjct: 3 RGIDFKRNNENGIDAHRFGELLMSSGIVLNDNVHWVXFPQXVTEFRVPLLKLLXVSGIXP 182
Query: 191 SRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVGN 239
F +++ F VYD+K+++KFCN L+GGL ++A L+V R VGN
Sbjct: 183 DTQVGFFNLINMYF-PTVYDIKHLMKFCNSLHGGLNKLAELLEVER-VGN 326
Score = 30.0 bits (66), Expect(2) = 2e-13
Identities = 16/35 (45%), Positives = 24/35 (67%)
Frame = +2
Query: 245 SDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
S+S LT F+K+KD +F + + +AGVL+GL V
Sbjct: 344 SNSWLTSCTFRKLKDNFF-SGSLEKYAGVLYGLGV 445
>AW597953
Length = 323
Score = 56.6 bits (135), Expect = 1e-08
Identities = 24/41 (58%), Positives = 32/41 (77%)
Frame = +3
Query: 9 ASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPG 49
A + I+IR+VW +NLE EF LIR+ V +PY++MDTEFPG
Sbjct: 138 AKSDSIQIREVWNDNLEEEFALIREIVDNYPYIAMDTEFPG 260
>TC214585 weakly similar to UP|PTRB_MORLA (Q59536) Protease II
(Oligopeptidase B) , partial (17%)
Length = 1249
Score = 30.0 bits (66), Expect = 1.2
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = +2
Query: 193 LEEFLSILTQIFGQNVYDMKYMI 215
L +FLS + Q QNVY+MKY++
Sbjct: 407 LSKFLSFIIQASPQNVYEMKYLV 475
>BE659930
Length = 109
Score = 28.5 bits (62), Expect = 3.6
Identities = 14/24 (58%), Positives = 14/24 (58%)
Frame = +3
Query: 73 YSFLKANVDNLNLIQLGLTLTDAN 96
Y L NVD L L QLGLT D N
Sbjct: 36 YQTLTXNVDMLKLXQLGLTFYDEN 107
>TC215592 similar to UP|Q9SSS9 (Q9SSS9) H+-transporting ATP synthase-like
protein, partial (87%)
Length = 961
Score = 27.7 bits (60), Expect = 6.1
Identities = 24/58 (41%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Frame = +2
Query: 166 VSWVTFHSSYDFGYLVKILTQNYLPSRLEEFLSI-LTQIFG--QNVYDMKYMIKFCNL 220
+SWV HS YD + + + LPS F+SI T IFG NVY + M +F NL
Sbjct: 746 LSWVISHSPYD----ISLPSVALLPSFCLFFVSISFTIIFGAKYNVY*IL*MNEFQNL 907
>TC234750 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY4, partial
(11%)
Length = 934
Score = 27.7 bits (60), Expect = 6.1
Identities = 18/59 (30%), Positives = 29/59 (48%)
Frame = +3
Query: 184 LTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCNLYGGLERVATKLKVSRAVGNSHQ 242
+TQ+ + L FL +F D KY ++F NL+ G+E ++ V + SHQ
Sbjct: 243 ITQSIACA*LYNFLDCFV*VFA----DAKY*LRFLNLFLGVEVCEGQISVEKIW*KSHQ 407
>TC218600
Length = 627
Score = 27.7 bits (60), Expect = 6.1
Identities = 18/51 (35%), Positives = 27/51 (52%)
Frame = +1
Query: 95 ANGNLPGDVAYSYIWEFNFKDFDVDRDLQNPDSIELLRRQGIDFKRNLIYG 145
A+G + G AY+Y W+ VD + S +L RRQ ++ R L+YG
Sbjct: 43 AHGVMVGRAAYNYPWQIL---GHVDAAIYGTPSCDLTRRQVLE--RYLVYG 180
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.140 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,752,452
Number of Sequences: 63676
Number of extensions: 150788
Number of successful extensions: 680
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 653
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 655
length of query: 281
length of database: 12,639,632
effective HSP length: 96
effective length of query: 185
effective length of database: 6,526,736
effective search space: 1207446160
effective search space used: 1207446160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146332.3


