

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.11 - phase: 0
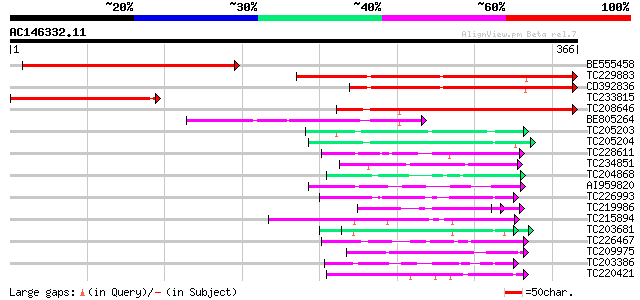
(366 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE555458 277 6e-75
TC229883 similar to GB|BAC20813.1|23617133|AP005183 transcriptio... 260 8e-70
CD392836 211 3e-55
TC233815 similar to UP|O64895 (O64895) Arabidopsis thaliana, par... 167 7e-42
TC208646 similar to UP|O64895 (O64895) Arabidopsis thaliana, par... 125 4e-29
BE805264 97 9e-21
TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall pr... 60 1e-09
TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein pr... 59 3e-09
TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxida... 53 2e-07
TC234851 weakly similar to UP|Q9DVW0 (Q9DVW0) PxORF73 peptide, p... 50 2e-06
TC204868 similar to UP|Q9LKY8 (Q9LKY8) Proline-rich protein, par... 49 3e-06
AI959820 similar to GP|14335118|gb| At1g30200/F12P21_1 {Arabidop... 49 3e-06
TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 deh... 49 3e-06
TC219986 weakly similar to UP|Q41187 (Q41187) Glycine-rich prote... 42 5e-06
TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase ... 48 8e-06
TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 48 8e-06
TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 47 1e-05
TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%) 47 1e-05
TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 47 1e-05
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 47 1e-05
>BE555458
Length = 426
Score = 277 bits (708), Expect = 6e-75
Identities = 136/140 (97%), Positives = 138/140 (98%)
Frame = +1
Query: 9 PNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRN 68
PNLDQ STKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRN
Sbjct: 7 PNLDQHSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRN 186
Query: 69 TQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVAN 128
TQPRFQFIVMNRRNTENLVENLLGDFEYE+Q+PYLLYRNAAQEVNGIWFYNARECEEVAN
Sbjct: 187 TQPRFQFIVMNRRNTENLVENLLGDFEYEVQIPYLLYRNAAQEVNGIWFYNARECEEVAN 366
Query: 129 LFSRILNAYAKVPPKLKVSS 148
LFSRILNAYAKVPPK KVSS
Sbjct: 367 LFSRILNAYAKVPPKSKVSS 426
>TC229883 similar to GB|BAC20813.1|23617133|AP005183 transcription
factor-like protein {Oryza sativa (japonica
cultivar-group);} , partial (15%)
Length = 1063
Score = 260 bits (664), Expect = 8e-70
Identities = 139/186 (74%), Positives = 148/186 (78%), Gaps = 5/186 (2%)
Frame = +3
Query: 186 PSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSG-PTHAATPVVPLPTLQIPSLPTST 244
PSF+NFFSAAM IGNTSN PITGQPYQSSA ISSS PTH ATP VP TLQIPSL ST
Sbjct: 3 PSFVNFFSAAMAIGNTSNTPITGQPYQSSAAISSSSVPTHTATPTVP--TLQIPSLSQST 176
Query: 245 PFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRP 304
P P DA ESI SS++ LVKPSFFVPP SSA MM+PPVSSS PTAPPLHP V RP
Sbjct: 177 PLIPPLDAVESICSSNRTAKLVKPSFFVPPPSSA-MMIPPVSSSTPTAPPLHPAVNVHRP 353
Query: 305 YGTPMLQPFPPPTPPPSLAPVSSPLPNP----AISREKVRDALLVLVQDNQFIDMVYKAL 360
YG P+LQPFPPP PP SLAP +SP +P ISREKVRDAL+VLVQDNQFIDMVY+AL
Sbjct: 354 YGAPLLQPFPPPNPPLSLAPAASPSSSPNYGSVISREKVRDALMVLVQDNQFIDMVYRAL 533
Query: 361 LNAHQS 366
LNAHQS
Sbjct: 534 LNAHQS 551
>CD392836
Length = 586
Score = 211 bits (538), Expect = 3e-55
Identities = 111/149 (74%), Positives = 121/149 (80%), Gaps = 2/149 (1%)
Frame = -1
Query: 220 SGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAA 279
S PT+AATP VP TLQIP+L STP PQ DA E I+SS++ LVKPSFFVPP SSA
Sbjct: 586 SVPTNAATPTVP--TLQIPTLSASTPLIPQLDAAELISSSNRTAKLVKPSFFVPPPSSA- 416
Query: 280 MMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPN--PAISRE 337
MM+PPVSSS P+APPLHP VQRPYG P+LQPFPPP PPPSLAP SP P+ P ISRE
Sbjct: 415 MMIPPVSSSTPSAPPLHPAVNVQRPYGAPLLQPFPPPNPPPSLAPAPSPSPSYGPVISRE 236
Query: 338 KVRDALLVLVQDNQFIDMVYKALLNAHQS 366
KVRDALLVLVQDNQFIDMVY+ALLNAHQS
Sbjct: 235 KVRDALLVLVQDNQFIDMVYRALLNAHQS 149
>TC233815 similar to UP|O64895 (O64895) Arabidopsis thaliana, partial (26%)
Length = 483
Score = 167 bits (423), Expect = 7e-42
Identities = 83/97 (85%), Positives = 91/97 (93%)
Frame = +3
Query: 1 MSQNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEG 60
MSQ KL PNLDQQSTK+LNLTVLQRIDPF+EEIL TAAHV+FY+FNI+ +QWSRKDVEG
Sbjct: 195 MSQTKKLTPNLDQQSTKVLNLTVLQRIDPFIEEILFTAAHVSFYDFNIESNQWSRKDVEG 374
Query: 61 SLFVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYE 97
SLFVVKRN+QPRFQFIVMNRRNT+NLVENLL DFEYE
Sbjct: 375 SLFVVKRNSQPRFQFIVMNRRNTDNLVENLL-DFEYE 482
>TC208646 similar to UP|O64895 (O64895) Arabidopsis thaliana, partial (12%)
Length = 980
Score = 125 bits (313), Expect = 4e-29
Identities = 72/158 (45%), Positives = 99/158 (62%), Gaps = 3/158 (1%)
Frame = +3
Query: 212 QSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQH--DAPESINSSSQATNLVKPS 269
Q SAT++S+ +T P +QIPS ST D+ E+ S +Q NLVKPS
Sbjct: 6 QHSATVTSAPSGLFSTA----PAVQIPSASHSTSSISGFPLDSLETAKSGNQIINLVKPS 173
Query: 270 FFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPL 329
F SS+++++PP+ SS+ + + + + R YGTP+LQPFPPP PP SL P+SS
Sbjct: 174 TFFASTSSSSLLVPPIYSSVQPSAAVQHSLNLPRQYGTPVLQPFPPPNPPLSLTPISSST 353
Query: 330 PN-PAISREKVRDALLVLVQDNQFIDMVYKALLNAHQS 366
PN P SR+ VRDALL LVQ++QFIDM+++ALL S
Sbjct: 354 PNRPVFSRDNVRDALLSLVQEDQFIDMMFQALLKVSHS 467
>BE805264
Length = 445
Score = 97.4 bits (241), Expect = 9e-21
Identities = 64/157 (40%), Positives = 83/157 (52%), Gaps = 2/157 (1%)
Frame = +2
Query: 115 IWFYNARECEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSS 174
IWFYN ECEEVANLF+RILNAY K PP SE EEL+ P + + PLE SS
Sbjct: 2 IWFYNPDECEEVANLFNRILNAYPKTPPTTIPPVNISESEELQ--PVSIIPESPLESSSG 175
Query: 175 ITSNVADVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPT 234
S + D +DP+F NFFS GN ++ + + ++ T + G A PT
Sbjct: 176 AASAI-DAVEDPAFTNFFSTLKVTGNYASNMENFRQHSATVTSAPGGLLSTA------PT 334
Query: 235 LQIPSLPTSTPFTPQH--DAPESINSSSQATNLVKPS 269
++IPS ST D+ E+ S +Q N VKPS
Sbjct: 335 VRIPSASHSTSSMSGFALDSLETAKSGNQVINRVKPS 445
>TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall protein,
partial (48%)
Length = 1076
Score = 60.5 bits (145), Expect = 1e-09
Identities = 47/148 (31%), Positives = 59/148 (39%), Gaps = 4/148 (2%)
Frame = +3
Query: 192 FSAAMGIGNTSNAPITGQ----PYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFT 247
F+ G+G + P T P S + SS P P P P+ P T P
Sbjct: 237 FTLVAGVGAQAQGPSTSPAPPTPQASPPPVQSSPPP---VPSSPPPSQSPPPASTPPPSP 407
Query: 248 PQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGT 307
P S SS + P+ PP SS PP SS P++PP P T
Sbjct: 408 LSSPPPSSPPPSSPPPSSPPPAS--PPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPAT 581
Query: 308 PMLQPFPPPTPPPSLAPVSSPLPNPAIS 335
P PP PPPSL P +PL +P +S
Sbjct: 582 P-----PPAVPPPSLTPTVTPLSSPPVS 650
>TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (44%)
Length = 996
Score = 58.9 bits (141), Expect = 3e-09
Identities = 45/149 (30%), Positives = 57/149 (38%), Gaps = 3/149 (2%)
Frame = +1
Query: 194 AAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAP 253
A +G S +P P S I SS P ++P P Q P ++ P P P
Sbjct: 163 AGVGAQGPSTSPAPPTPQSSPPPIQSSPPPVPSSP----PPAQSPPPASTPPPAPLSSPP 330
Query: 254 ESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPF 313
+ S P PP S PP S + PP P P+ P P
Sbjct: 331 PASPPPSSPPPASPPPASPPPASPPPASPPPASPPPASPPPATPPPASPPPFSPPPATP- 507
Query: 314 PPPTPPPSLAPV---SSPLPNPAISREKV 339
PP TPPP+L P S P +PA + KV
Sbjct: 508 PPATPPPALTPTPLSSPPATSPAPAPAKV 594
>TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26a
{Arabidopsis thaliana;} , partial (49%)
Length = 731
Score = 53.1 bits (126), Expect = 2e-07
Identities = 44/140 (31%), Positives = 63/140 (44%), Gaps = 9/140 (6%)
Frame = +3
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQ 261
S AP P +S++ ++S PT A TP P P P++TP T +P + ++SS
Sbjct: 165 SPAPPPPPPRSASSSTTASSPT-AVTPPSSSPPRPSPG-PSATP-TSTSPSPATPSTSSS 335
Query: 262 ATNLVKPSFFVPPLSSAAMMM---------PPVSSSIPTAPPLHPTGTVQRPYGTPMLQP 312
A PPLSS A PP +SS +A P P+ + P P
Sbjct: 336 APR--------PPLSSPAPTPSPAPTFSPPPPATSSPCSAAPSSPSSSAAATAEPPSPPP 491
Query: 313 FPPPTPPPSLAPVSSPLPNP 332
+PP +PPP + SP +P
Sbjct: 492 YPPTSPPPPCPSLKSPNSSP 551
Score = 39.7 bits (91), Expect = 0.002
Identities = 38/136 (27%), Positives = 55/136 (39%), Gaps = 2/136 (1%)
Frame = +3
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQ 261
S P P+ S+ T+ S TP S + +P P P S+S
Sbjct: 69 SAPPTRDSPWTSTRTLVRSSARS*GTP----------SQASRSPAPP----PPPPRSASS 206
Query: 262 ATNLVKPSFFVPPLSSAAMMMP-PVSSSIPTAP-PLHPTGTVQRPYGTPMLQPFPPPTPP 319
+T P+ PP SS P P ++ T+P P P+ + P P+ P P P+P
Sbjct: 207 STTASSPTAVTPPSSSPPRPSPGPSATPTSTSPSPATPSTSSSAPR-PPLSSPAPTPSPA 383
Query: 320 PSLAPVSSPLPNPAIS 335
P+ +P P PA S
Sbjct: 384 PTFSP-----PPPATS 416
>TC234851 weakly similar to UP|Q9DVW0 (Q9DVW0) PxORF73 peptide, partial (30%)
Length = 466
Score = 50.1 bits (118), Expect = 2e-06
Identities = 37/124 (29%), Positives = 56/124 (44%), Gaps = 6/124 (4%)
Frame = +3
Query: 214 SATISSSGPTHAATPVV------PLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVK 267
+AT S P+ ++ P + P P+ PS P S+ + P H +P + S AT+
Sbjct: 99 AATKWCSHPSCSSPPFLYRPW*FPNPSPPTPSPPKSSTWRPSHTSPPHV--SRPATSAKN 272
Query: 268 PSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSS 327
P+ P S++ P +S P+ PP +P T TP P PPP P + S
Sbjct: 273 PATKPSPSSASRPSSPSPPTSSPSPPP-YPPSTPPSSLSTPPPPPPPPPPPSATTPRAFS 449
Query: 328 PLPN 331
P P+
Sbjct: 450 PPPS 461
>TC204868 similar to UP|Q9LKY8 (Q9LKY8) Proline-rich protein, partial (91%)
Length = 865
Score = 49.3 bits (116), Expect = 3e-06
Identities = 43/129 (33%), Positives = 46/129 (35%)
Frame = +2
Query: 205 PITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATN 264
P T P S + PT TP P PT P P TP TP+ P +
Sbjct: 128 PPTPTPSPSPKPTPPTPPTPKPTPPTPKPT---PPTPKPTPPTPKPTPPTPTPT------ 280
Query: 265 LVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAP 324
PP A PP S PT PP PT P TP P PPTP P
Sbjct: 281 --------PPTPKAGC--PPPSPPKPT-PPTPPTPKPTPPTPTPTPTPPTPPTPKAGCPP 427
Query: 325 VSSPLPNPA 333
S P P A
Sbjct: 428 PSPPSPPKA 454
>AI959820 similar to GP|14335118|gb| At1g30200/F12P21_1 {Arabidopsis
thaliana}, partial (19%)
Length = 342
Score = 49.3 bits (116), Expect = 3e-06
Identities = 41/140 (29%), Positives = 58/140 (41%)
Frame = +2
Query: 194 AAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAP 253
AA ++ P + + SS+ ++S PT TP+ P P P+ P+ T
Sbjct: 23 AAAASSRAASTPSSRRSKTSSSASTASSPT--TTPLPPPPPPTSPAAPSGT--------- 169
Query: 254 ESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPF 313
S +SS+++ N KPS VSSS P APP P+ +P
Sbjct: 170 SSASSSAES*NPSKPS---------------VSSSAPNAPPPPPS------------RPL 268
Query: 314 PPPTPPPSLAPVSSPLPNPA 333
PPP PPP + P PA
Sbjct: 269 PPPPPPPPSPSEPTTTPTPA 328
>TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 dehydratase
(GDP-D-mannose dehydratase) (GMD) , partial (94%)
Length = 1388
Score = 49.3 bits (116), Expect = 3e-06
Identities = 39/128 (30%), Positives = 60/128 (46%)
Frame = +2
Query: 201 TSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSS 260
+S PI+ P S+A + S PT + T + L + PS P+ P TP +P + ++SS
Sbjct: 311 SSTTPISPTPPPSAAGSTPSSPTRSTTSPLSLTS---PS-PSRYPTTPPTSSPPAPSASS 478
Query: 261 QATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPP 320
+ + P PP + P S++ APP ++R P P PPTPPP
Sbjct: 479 RPSVPTSP----PPAA-------PTSATTKPAPPRCSAPPLRRSPKPPPSTP-APPTPPP 622
Query: 321 SLAPVSSP 328
+ P +P
Sbjct: 623 NAPPTGTP 646
Score = 30.0 bits (66), Expect = 1.7
Identities = 26/89 (29%), Positives = 37/89 (41%), Gaps = 1/89 (1%)
Frame = +2
Query: 252 APESINSSSQATNLVKPSFFVPPL-SSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPML 310
AP + S+ T++ P+ P SS + P S + P PT R +P+
Sbjct: 236 APRTSTRSASTTSMWIPTTPTRPA*SSTTPISPTPPPSAAGSTPSSPT----RSTTSPLS 403
Query: 311 QPFPPPTPPPSLAPVSSPLPNPAISREKV 339
P P+ P+ P SSP A SR V
Sbjct: 404 LTSPSPSRYPTTPPTSSPPAPSASSRPSV 490
>TC219986 weakly similar to UP|Q41187 (Q41187) Glycine-rich protein
(Fragment), partial (47%)
Length = 584
Score = 41.6 bits (96), Expect(2) = 5e-06
Identities = 30/96 (31%), Positives = 39/96 (40%)
Frame = -3
Query: 225 AATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPP 284
A+T + P T IP + + P TP + P L P PP+ M PP
Sbjct: 318 ASTCLKPPMTPPIPPIAPTPPPTPPNPIP-----------L*MP----PPIPIPL*MPPP 184
Query: 285 VSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPP 320
+ P PP PT + P P +P PP PPP
Sbjct: 183 IPPPRPLPPPSPPTPPLPAPPPYPPPKPLSPPAPPP 76
Score = 31.2 bits (69), Expect = 0.77
Identities = 19/52 (36%), Positives = 21/52 (39%)
Frame = -3
Query: 281 MMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNP 332
M PP+ PT PP P P P P P PPP P P P+P
Sbjct: 294 MTPPIPPIAPTPPPTPPNPI---PL*MPPPIPIPL*MPPPIPPPRPLPPPSP 148
Score = 26.2 bits (56), Expect(2) = 5e-06
Identities = 10/21 (47%), Positives = 12/21 (56%)
Frame = -1
Query: 312 PFPPPTPPPSLAPVSSPLPNP 332
P PPP PP S P + P+ P
Sbjct: 65 PKPPPAPPTSPIPPTPPMDPP 3
>TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase , complete
Length = 1675
Score = 47.8 bits (112), Expect = 8e-06
Identities = 50/170 (29%), Positives = 72/170 (41%), Gaps = 8/170 (4%)
Frame = +1
Query: 168 PLEPSSSITS-NVADVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSG---PT 223
PL P S T+ + + P + + +++ TS++ T P S+A S+S P+
Sbjct: 244 PLPPRPSATTRSPSSAPATWEWPSRRPSSLRTSPTSSSSSTPTPTSSAARCSTSSTPPPS 423
Query: 224 HAATPVVPLPTLQIPSLPT--STPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMM 281
A P PT P PT S+P P ++SS+ T+ F PLSS +
Sbjct: 424 SPAPRSTPPPTPPSPPAPTSASSPPAPARSPASHASTSSRGTSPSSAPLF--PLSS---V 588
Query: 282 MPP--VSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPL 329
PP +SSS PT PT P P P PT + SSP+
Sbjct: 589 TPPTQLSSSFPTPSTSSPTSRGSSPASPPTASSAPAPTWTLPASVSSSPI 738
Score = 37.7 bits (86), Expect = 0.008
Identities = 42/125 (33%), Positives = 53/125 (41%), Gaps = 1/125 (0%)
Frame = +1
Query: 219 SSGP-THAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSS 277
SSGP T P P T + PS +T P P S+ +S +++ P+ P SS
Sbjct: 220 SSGP*TTPPLPPRPSATTRSPSSAPATWEWPSR-RPSSLRTSPTSSSSSTPT----PTSS 384
Query: 278 AAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISRE 337
AA S+ T PP P P TP PPTPP AP S+ P PA +R
Sbjct: 385 AARC-----STSSTPPPSSPA-----PRSTP------PPTPPSPPAPTSASSP-PAPARS 513
Query: 338 KVRDA 342
A
Sbjct: 514 PASHA 528
>TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 913
Score = 47.8 bits (112), Expect = 8e-06
Identities = 35/140 (25%), Positives = 53/140 (37%), Gaps = 2/140 (1%)
Frame = +3
Query: 201 TSNAPITGQPYQSSATISSS--GPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINS 258
TS T P+ S AT S P A+P P P+ T+TP +
Sbjct: 99 TSPPAATTTPFASPATAPSKPKSPAPVASPTSSSPPASSPNAATATPPASSPTVASPPSK 278
Query: 259 SSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTP 318
++ + P PP ++ PP + + + P P + P P TP
Sbjct: 279 AAAPAPVATPPAATPPAATPPAATPPAVTPVSSPPAPVPVSSPPAPVPVSSPPALAPTTP 458
Query: 319 PPSLAPVSSPLPNPAISREK 338
P +AP S+ +P PA +K
Sbjct: 459 APVVAP-SAEVPAPAPKSKK 515
Score = 44.3 bits (103), Expect = 9e-05
Identities = 37/126 (29%), Positives = 49/126 (38%), Gaps = 12/126 (9%)
Frame = +3
Query: 215 ATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAP-ESINSSSQATNLVKPSFFVP 273
A + P + T P T + P + P P+ AP S SSS + + P
Sbjct: 60 AAVGGQSPAASPTTSPPAATTTPFASPATAPSKPKSPAPVASPTSSSPPASSPNAATATP 239
Query: 274 PLSSAAMMMPP--------VSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTP---PPSL 322
P SS + PP V++ PP P TP+ P P P P PP+
Sbjct: 240 PASSPTVASPPSKAAAPAPVATPPAATPPAATPPAATPPAVTPVSSP-PAPVPVSSPPAP 416
Query: 323 APVSSP 328
PVSSP
Sbjct: 417 VPVSSP 434
Score = 28.5 bits (62), Expect = 5.0
Identities = 15/45 (33%), Positives = 22/45 (48%)
Frame = +3
Query: 286 SSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLP 330
++S T+PP T P+ +P P P +P P +P SS P
Sbjct: 84 AASPTTSPPAATT----TPFASPATAPSKPKSPAPVASPTSSSPP 206
>TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 1017
Score = 47.4 bits (111), Expect = 1e-05
Identities = 46/135 (34%), Positives = 58/135 (42%), Gaps = 1/135 (0%)
Frame = +3
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPT-STPFTPQHDAPESINSSS 260
+ AP P +AT SSG A T LP + S PT STP T +S
Sbjct: 57 ATAPANSPPTTPAATSPSSGTPLAVT----LPPTVVASPPTTSTPPT-----------TS 191
Query: 261 QATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPP 320
Q +V P P++ A + P SS P APP T Q P +P++ P PP PP
Sbjct: 192 QPPAIVAPK--PAPVTPPAPKVAPASS--PKAPPPQ-TPQPQPPKVSPVVTPTSPPPIPP 356
Query: 321 SLAPVSSPLPNPAIS 335
PLP P I+
Sbjct: 357 PPVSTPPPLPPPKIA 401
Score = 39.7 bits (91), Expect = 0.002
Identities = 38/155 (24%), Positives = 55/155 (34%), Gaps = 25/155 (16%)
Frame = +3
Query: 202 SNAPITGQPYQSSA--TISSSGPTHAATP--------VVPLPTLQIPSLPTSTPFT---- 247
+ +P +G P + T+ +S PT + P V P P P P P +
Sbjct: 96 ATSPSSGTPLAVTLPPTVVASPPTTSTPPTTSQPPAIVAPKPAPVTPPAPKVAPASSPKA 275
Query: 248 --PQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQ--- 302
PQ P+ S T P PP+S+ + PP + P P P
Sbjct: 276 PPPQTPQPQPPKVSPVVTPTSPPPIPPPPVSTPPPLPPPKIAPTPAKTPPAPAPAKATPA 455
Query: 303 ------RPYGTPMLQPFPPPTPPPSLAPVSSPLPN 331
+P TP P PP P + V +P P+
Sbjct: 456 PAPAPTKPAPTPAPVPPPPTLAPTPVVEVPAPAPS 560
>TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%)
Length = 537
Score = 47.4 bits (111), Expect = 1e-05
Identities = 39/119 (32%), Positives = 51/119 (42%), Gaps = 1/119 (0%)
Frame = +1
Query: 218 SSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSS 277
S S P P P P+ Q P PT P TP + + + PS PP +
Sbjct: 100 SYSPPNVPTPPKTPSPSSQPPFTPTP-PKTPSPTSQPPYIPTPPSPISQPPSIATPPNTL 276
Query: 278 AAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPS-LAPVSSPLPNPAIS 335
+ PP S+IP + L P+ P P PT PPS LAP +SP P+P+ S
Sbjct: 277 SPTSQPPYPSTIPPSTSLSPSYP-------PSPSPSSAPTYPPSYLAPATSP-PSPSTS 429
Score = 38.1 bits (87), Expect = 0.006
Identities = 34/105 (32%), Positives = 45/105 (42%), Gaps = 4/105 (3%)
Frame = +1
Query: 204 APITGQPYQSSATISSSGPTHAATPVVPL-PTLQIP---SLPTSTPFTPQHDAPESINSS 259
+P + PY + S P ATP L PT Q P ++P ST +P + P S + S
Sbjct: 190 SPTSQPPYIPTPPSPISQPPSIATPPNTLSPTSQPPYPSTIPPSTSLSPSY--PPSPSPS 363
Query: 260 SQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRP 304
S T PS+ P S PP S+ T P + P T P
Sbjct: 364 SAPT--YPPSYLAPATS------PPSPSTSATTPTISPAATTPSP 474
Score = 38.1 bits (87), Expect = 0.006
Identities = 21/59 (35%), Positives = 28/59 (46%), Gaps = 1/59 (1%)
Frame = +1
Query: 273 PPLSSAAMMMPPVSSSIPTAPPLHPT-GTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLP 330
PP + + + PP++ S PP PT P +P P PP TP PS P +P P
Sbjct: 1 PPKTPSPIYAPPIAPSPGNHPPYVPTPPKTPSPSYSPPNVPTPPKTPSPSSQPPFTPTP 177
Score = 36.6 bits (83), Expect = 0.018
Identities = 46/158 (29%), Positives = 62/158 (39%), Gaps = 4/158 (2%)
Frame = +1
Query: 168 PLEPSSSITS-NVADVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAA 226
P PS S + NV P PS +S P T P ++ + S P +
Sbjct: 82 PKTPSPSYSPPNVPTPPKTPS------------PSSQPPFTPTPPKTPSPTSQ--PPYIP 219
Query: 227 TPVVPLPTLQIPSL---PTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMP 283
TP P P Q PS+ P + T Q P +I S+ + PS+ P S+A P
Sbjct: 220 TP--PSPISQPPSIATPPNTLSPTSQPPYPSTIPPSTS----LSPSYPPSPSPSSAPTYP 381
Query: 284 PVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPS 321
P + T+PP T TP + P TP PS
Sbjct: 382 PSYLAPATSPPSPSTSAT-----TPTISP-AATTPSPS 477
Score = 34.3 bits (77), Expect = 0.091
Identities = 22/64 (34%), Positives = 31/64 (48%)
Frame = +1
Query: 273 PPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNP 332
PP + + PP ++PT PP P+ + Q P+ P PP TP P+ P P P
Sbjct: 79 PPKTPSPSYSPP---NVPT-PPKTPSPSSQPPF-----TPTPPKTPSPTSQPPYIPTPPS 231
Query: 333 AISR 336
IS+
Sbjct: 232 PISQ 243
Score = 28.1 bits (61), Expect = 6.5
Identities = 41/144 (28%), Positives = 58/144 (39%), Gaps = 15/144 (10%)
Frame = +1
Query: 141 PPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAMGIGN 200
PPK S++ F PT P PS TS +P PS I+ + N
Sbjct: 127 PPKTPSPSSQPPF-----TPTP-----PKTPSP--TSQPPYIPTPPSPISQPPSIATPPN 270
Query: 201 TSNAPITGQPYQSSATIS-------------SSGPTHAATPVVPLPTLQIPSLPTSTP-F 246
T + P + PY S+ S SS PT+ + + P + PS +TP
Sbjct: 271 TLS-PTSQPPYPSTIPPSTSLSPSYPPSPSPSSAPTYPPSYLAPATSPPSPSTSATTPTI 447
Query: 247 TPQHDAPE-SINSSSQATNLVKPS 269
+P P S +SSS A+N P+
Sbjct: 448 SPAATTPSPSSSSSSGASNETTPA 519
>TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 649
Score = 47.0 bits (110), Expect = 1e-05
Identities = 42/126 (33%), Positives = 57/126 (44%), Gaps = 1/126 (0%)
Frame = +1
Query: 204 APITGQPYQSSATISSSGPTHAATPVVPLPTLQIP-SLPTSTPFTPQHDAPESINSSSQA 262
A + GQ ++ T SS+ P A P P P P + PTS+P P S +++ A
Sbjct: 55 AAVGGQSPAAAPTTSSASP--ATAPSKPKPKSPAPVASPTSSP-------PASSPNAATA 207
Query: 263 TNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSL 322
T PP+SS + PP ++ P AP P P TP PP PP++
Sbjct: 208 T---------PPVSSPTVASPPSKAAAP-APATTP------PVATPPAAT-PPAATPPAV 336
Query: 323 APVSSP 328
PVSSP
Sbjct: 337 TPVSSP 354
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 47.0 bits (110), Expect = 1e-05
Identities = 45/148 (30%), Positives = 62/148 (41%), Gaps = 17/148 (11%)
Frame = +3
Query: 205 PITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESIN----SSS 260
P T P+++ A S +ATP P T + PT+ P +P P S + S
Sbjct: 204 PSTSPPHRTPAPSPPSTMPPSATPPSPSATSSPSASPTAPPTSPSPSPPSSPSPPSAPSP 383
Query: 261 QATNLVKPSFFVP---------PLSSAAMMMP----PVSSSIPTAPPLHPTGTVQRPYGT 307
+T PS P P S+A+ P P S + PT+P PT T + P +
Sbjct: 384 SSTAPSPPSASPPTSPSPSPPSPASTASANSPASGSPTSRTSPTSP--SPTSTSKPPAPS 557
Query: 308 PMLQPFPPPTPPPSLAPVSSPLPNPAIS 335
P P PS P+ SP P+PA S
Sbjct: 558 SSSPA*PSSKPSPSPTPI-SPDPSPATS 638
Score = 35.8 bits (81), Expect = 0.031
Identities = 28/94 (29%), Positives = 38/94 (39%), Gaps = 2/94 (2%)
Frame = +3
Query: 241 PTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGT 300
P + +P H P S+ + PS SA+ PP S S +PP P
Sbjct: 195 PLNPSTSPPHRTPAPSPPSTMPPSATPPSPSATSSPSASPTAPPTSPS--PSPPSSP--- 359
Query: 301 VQRPYGTPMLQPFPPPT--PPPSLAPVSSPLPNP 332
+P P P T PPS +P +SP P+P
Sbjct: 360 ------SPPSAPSPSSTAPSPPSASPPTSPSPSP 443
Score = 35.8 bits (81), Expect = 0.031
Identities = 41/164 (25%), Positives = 71/164 (43%), Gaps = 1/164 (0%)
Frame = +3
Query: 168 PLEPSSSITSNVADVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAAT 227
P PS+S ++ + P P+ S A ++N+P +G P ++ S + P+ +T
Sbjct: 396 PSPPSASPPTSPSPSPPSPA-----STA-----SANSPASGSP---TSRTSPTSPSPTST 536
Query: 228 PVVPLPTLQIPSLPTSTPF-TPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVS 286
P P+ P+ P+S P +P +P+ ++S T+ S P +S A + P
Sbjct: 537 SKPPAPSSSSPA*PSSKPSPSPTPISPDPSPATSTPTSHTSIS---PITASKATFLLP*L 707
Query: 287 SSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLP 330
S ++ + P Y P + F T P P SP+P
Sbjct: 708 CSTASSF*ISPLTPSPEKYLLPSVT*FHSKTSPS--PPTPSPVP 833
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.131 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,237,097
Number of Sequences: 63676
Number of extensions: 324653
Number of successful extensions: 11970
Number of sequences better than 10.0: 1247
Number of HSP's better than 10.0 without gapping: 5265
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8049
length of query: 366
length of database: 12,639,632
effective HSP length: 99
effective length of query: 267
effective length of database: 6,335,708
effective search space: 1691634036
effective search space used: 1691634036
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146332.11


