

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146331.8 + phase: 2 /pseudo
(279 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
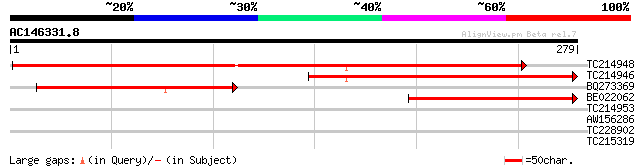
Score E
Sequences producing significant alignments: (bits) Value
TC214948 weakly similar to UP|Q9ZVV1 (Q9ZVV1) T5A14.4 protein, p... 393 e-110
TC214946 homologue to UP|Q6TEC7 (Q6TEC7) Defender against progra... 210 5e-55
BQ273369 145 2e-35
BE022062 similar to GP|20453070|gb| AT5g66920/MUD21_18 {Arabidop... 125 3e-29
TC214953 35 0.029
AW156286 35 0.038
TC228902 similar to UP|Q9XHD3 (Q9XHD3) PHAP2B protein, partial (... 29 2.1
TC215319 similar to UP|Q6Z1N4 (Q6Z1N4) Type 1 membrane protein-l... 28 3.5
>TC214948 weakly similar to UP|Q9ZVV1 (Q9ZVV1) T5A14.4 protein, partial (58%)
Length = 1024
Score = 393 bits (1009), Expect = e-110
Identities = 210/254 (82%), Positives = 220/254 (85%), Gaps = 1/254 (0%)
Frame = +1
Query: 2 SENGLKESVNYQVISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDLS 61
SENGLKESVNYQVIS KDLAKSV SEAGWSNFLCKGKK +PLDLALLFVG ELQSSDL+
Sbjct: 265 SENGLKESVNYQVISPKDLAKSVFSEAGWSNFLCKGKKLHEPLDLALLFVGRELQSSDLN 444
Query: 62 LNKHADSALSYLLKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVAF 121
+NKHA+SAL LLK SF RSNTS+AFPYVS SEDV LE+SLVSGFAEACG D+GIGNVAF
Sbjct: 445 MNKHANSALLDLLKISFARSNTSVAFPYVSTSEDVLLENSLVSGFAEACG-DMGIGNVAF 621
Query: 122 LGSCSMGNGNREETAALQSVQAYLTKRKEESHKGKTDLVVFCN-GPQASKNVDSTKSEGE 180
GSCSM N EE L SVQ YL KR EESH+GKTDLVVFCN G QA KNVD T+SEGE
Sbjct: 622 HGSCSMDGANHEEITTLHSVQDYLIKRMEESHRGKTDLVVFCNEGSQAVKNVDKTQSEGE 801
Query: 181 VLSELISSVEESGAKYAVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTNSTACDGVCQ 240
VLS LISSVEESGAKYAVLYVSD SRSIQYPSYRDLQRFLAEST GN STNST CD VCQ
Sbjct: 802 VLSNLISSVEESGAKYAVLYVSDPSRSIQYPSYRDLQRFLAESTAGNESTNSTICDEVCQ 981
Query: 241 LKSSLLEGLLVGIV 254
LKSSLLEG+LVGIV
Sbjct: 982 LKSSLLEGILVGIV 1023
>TC214946 homologue to UP|Q6TEC7 (Q6TEC7) Defender against programmed cell
death, partial (12%)
Length = 1085
Score = 210 bits (535), Expect = 5e-55
Identities = 110/133 (82%), Positives = 117/133 (87%), Gaps = 1/133 (0%)
Frame = +3
Query: 148 RKEESHKGKTDLVVFCN-GPQASKNVDSTKSEGEVLSELISSVEESGAKYAVLYVSDISR 206
R ES +GKTD VVF N G QA +NVD T+SEGEVLS LI+SVEESGAKYA+LYVSD SR
Sbjct: 3 RXXESXRGKTDXVVFXNEGSQALENVDRTQSEGEVLSNLINSVEESGAKYAILYVSDPSR 182
Query: 207 SIQYPSYRDLQRFLAESTTGNGSTNSTACDGVCQLKSSLLEGLLVGIVLLIILISGLCCM 266
SIQYPSYRDLQRFLAEST GN STNST CD VCQLKSSLLEG+LVGIVLLIILISGLCCM
Sbjct: 183 SIQYPSYRDLQRFLAESTAGNESTNSTICDEVCQLKSSLLEGILVGIVLLIILISGLCCM 362
Query: 267 MGIDSPTRFEAPQ 279
MGID+PTRFE PQ
Sbjct: 363 MGIDTPTRFETPQ 401
>BQ273369
Length = 421
Score = 145 bits (366), Expect = 2e-35
Identities = 84/139 (60%), Positives = 91/139 (65%), Gaps = 40/139 (28%)
Frame = +1
Query: 14 VISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDLSLNKHADSALSYL 73
VIS KDLAKS+LSEAGWSNFLCKGKKF +PLDLALLFVG ELQSSDLS+NKHA+SAL L
Sbjct: 1 VISPKDLAKSILSEAGWSNFLCKGKKFHEPLDLALLFVGRELQSSDLSMNKHANSALLDL 180
Query: 74 LK----------------------------------------DSFVRSNTSMAFPYVSAS 93
LK +SF RSNTS+AFPYVS S
Sbjct: 181 LKVSKL*HAVCQLSMSSNPDLRFHKELCFFFFTVILFHCHEQNSFARSNTSVAFPYVSTS 360
Query: 94 EDVNLEDSLVSGFAEACGD 112
EDV LE+SLVSGFAEACGD
Sbjct: 361 EDVLLENSLVSGFAEACGD 417
>BE022062 similar to GP|20453070|gb| AT5g66920/MUD21_18 {Arabidopsis
thaliana}, partial (7%)
Length = 355
Score = 125 bits (313), Expect = 3e-29
Identities = 62/83 (74%), Positives = 70/83 (83%)
Frame = +3
Query: 197 AVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTNSTACDGVCQLKSSLLEGLLVGIVLL 256
A+ + + +S+S+ Y DLQRFLAEST GN STNST CD VCQLKSSLLEG+LVGIVLL
Sbjct: 102 AISWATVLSQSVGCTYYTDLQRFLAESTAGNESTNSTICDEVCQLKSSLLEGILVGIVLL 281
Query: 257 IILISGLCCMMGIDSPTRFEAPQ 279
IILISGLCCMMGID+PTRFE PQ
Sbjct: 282 IILISGLCCMMGIDTPTRFETPQ 350
>TC214953
Length = 992
Score = 35.4 bits (80), Expect = 0.029
Identities = 17/23 (73%), Positives = 19/23 (81%)
Frame = +1
Query: 229 STNSTACDGVCQLKSSLLEGLLV 251
STN T D VC+LKSSLLEG+LV
Sbjct: 4 STNFTISDEVCKLKSSLLEGILV 72
>AW156286
Length = 380
Score = 35.0 bits (79), Expect = 0.038
Identities = 19/50 (38%), Positives = 28/50 (56%)
Frame = +3
Query: 229 STNSTACDGVCQLKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAP 278
S+NS CD C L+ + G+ G+ L+I +GL M GI +PT + P
Sbjct: 9 SSNSAICDV*CHLQ*DVGMGMPGGVDWLLIYTAGLSWMTGIATPTIYVTP 158
>TC228902 similar to UP|Q9XHD3 (Q9XHD3) PHAP2B protein, partial (36%)
Length = 1242
Score = 29.3 bits (64), Expect = 2.1
Identities = 23/79 (29%), Positives = 38/79 (47%)
Frame = +2
Query: 151 ESHKGKTDLVVFCNGPQASKNVDSTKSEGEVLSELISSVEESGAKYAVLYVSDISRSIQY 210
E+ + + CNG +A N + + EGEV+S+ S E++ D++ +
Sbjct: 320 EAARAYDKAAIKCNGREAVTNFEPSFYEGEVISQ--SDNEDNKQSL------DLNLGLAP 475
Query: 211 PSYRDLQRFLAESTTGNGS 229
PSY D Q ++T NGS
Sbjct: 476 PSYSDGQ---IKNTPSNGS 523
>TC215319 similar to UP|Q6Z1N4 (Q6Z1N4) Type 1 membrane protein-like, partial
(12%)
Length = 665
Score = 28.5 bits (62), Expect = 3.5
Identities = 24/120 (20%), Positives = 51/120 (42%)
Frame = +3
Query: 150 EESHKGKTDLVVFCNGPQASKNVDSTKSEGEVLSELISSVEESGAKYAVLYVSDISRSIQ 209
+E++KG+ +++C+ +++ +ESG K+ V++
Sbjct: 33 QEAYKGQIXXIIYCH---------------------VATTQESGKKFDVIFTP------- 128
Query: 210 YPSYRDLQRFLAESTTGNGSTNSTACDGVCQLKSSLLEGLLVGIVLLIILISGLCCMMGI 269
+ R+LAE+ N + V ++ +L + GI+LLI G+C +M +
Sbjct: 129 ---HHPYARWLAETNAVNATLPE-----VVLVRRTL--AWVTGIILLISTFMGICYLMNM 278
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,550,078
Number of Sequences: 63676
Number of extensions: 126588
Number of successful extensions: 484
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 480
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 481
length of query: 279
length of database: 12,639,632
effective HSP length: 96
effective length of query: 183
effective length of database: 6,526,736
effective search space: 1194392688
effective search space used: 1194392688
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146331.8


