

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146331.7 - phase: 0
(377 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
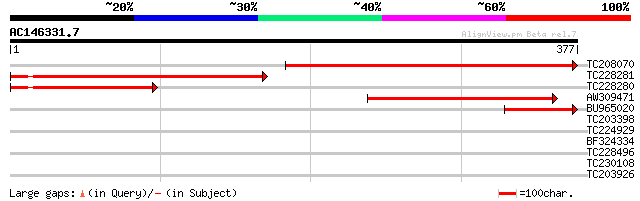
Score E
Sequences producing significant alignments: (bits) Value
TC208070 homologue to UP|BIOB_ARATH (P54967) Biotin synthase (B... 358 2e-99
TC228281 similar to UP|BIOB_ARATH (P54967) Biotin synthase (Bio... 291 4e-79
TC228280 similar to UP|BIOB_ARATH (P54967) Biotin synthase (Bio... 154 8e-38
AW309471 137 6e-33
BU965020 81 9e-16
TC203398 weakly similar to UP|Q7XYK7 (Q7XYK7) Ribosomal protein ... 31 0.80
TC224929 homologue to UP|Q6RYC4 (Q6RYC4) 60S ribosomal protein L... 30 1.4
BF324334 30 1.8
TC228496 29 4.0
TC230108 similar to UP|CNX2_ARATH (Q39055) Molybdopterin biosynt... 28 6.8
TC203926 weakly similar to PIR|T06696|T06696 lipase homolog T29H... 28 8.9
>TC208070 homologue to UP|BIOB_ARATH (P54967) Biotin synthase (Biotin
synthetase) , partial (46%)
Length = 841
Score = 358 bits (919), Expect = 2e-99
Identities = 179/194 (92%), Positives = 187/194 (96%)
Frame = +2
Query: 184 LKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGEAEDD 243
LKKAGLTAYNHNLDTSREYYPNIITTRTYDERL+TLEFVRDAGINVCSGGIIGLGEAE+D
Sbjct: 2 LKKAGLTAYNHNLDTSREYYPNIITTRTYDERLQTLEFVRDAGINVCSGGIIGLGEAEED 181
Query: 244 RVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKAMVRL 303
RVGLLHTLSTLPTHPESVPINAL+AVKGTPL+DQKPVEIWEMIRMIATARI MPKAMVRL
Sbjct: 182 RVGLLHTLSTLPTHPESVPINALVAVKGTPLEDQKPVEIWEMIRMIATARIIMPKAMVRL 361
Query: 304 SAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLPKAPSLDDDE 363
SAGRVRFS+PEQALCFLAGANSIF GEKLLTT NNDFDADQLMFKVLGLLPKAPSL + E
Sbjct: 362 SAGRVRFSMPEQALCFLAGANSIFTGEKLLTTPNNDFDADQLMFKVLGLLPKAPSLHEGE 541
Query: 364 TNEAENYKEAASSS 377
T+ E+YKEAASSS
Sbjct: 542 TSVTEDYKEAASSS 583
>TC228281 similar to UP|BIOB_ARATH (P54967) Biotin synthase (Biotin
synthetase) , partial (39%)
Length = 677
Score = 291 bits (744), Expect = 4e-79
Identities = 136/171 (79%), Positives = 155/171 (90%)
Frame = +2
Query: 1 MFWLRPILRSQSRSSIWVLQHCNSFSTSSAAAIQAEKTIQNGPRNDWTKDEVKSIYDSPI 60
MF RPI R S +W L ++S++SAAAIQAE+TI+ GPRNDW++D+VKSIYDSPI
Sbjct: 173 MFLARPIFRGPS---LWALHSSYAYSSASAAAIQAERTIKEGPRNDWSRDQVKSIYDSPI 343
Query: 61 LDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLNKDA 120
LDLLFHGAQVHRHAHNFREVQQCTLLS+KTGGCSEDCSYCPQSS+YDTG+KGQ+L+NK+A
Sbjct: 344 LDLLFHGAQVHRHAHNFREVQQCTLLSIKTGGCSEDCSYCPQSSKYDTGVKGQRLMNKEA 523
Query: 121 VLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIKGMGMEVCC 171
VLQAA KAKEAGSTRFCMGAAWRDT+GRKTNFNQILEYVK+I+ MGMEVCC
Sbjct: 524 VLQAAKKAKEAGSTRFCMGAAWRDTLGRKTNFNQILEYVKDIRDMGMEVCC 676
>TC228280 similar to UP|BIOB_ARATH (P54967) Biotin synthase (Biotin
synthetase) , partial (20%)
Length = 510
Score = 154 bits (388), Expect = 8e-38
Identities = 72/98 (73%), Positives = 84/98 (85%)
Frame = +1
Query: 1 MFWLRPILRSQSRSSIWVLQHCNSFSTSSAAAIQAEKTIQNGPRNDWTKDEVKSIYDSPI 60
MF RPI R+ S +W L ++S++SAAAIQAE+ I+ GPRNDW++D+VKSIYDSPI
Sbjct: 226 MFLARPIFRAPS---LWALHSSYAYSSASAAAIQAERAIKEGPRNDWSRDQVKSIYDSPI 396
Query: 61 LDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCS 98
LDLLFHGAQVHRHAHNFREVQQCTLLS+KTGGCSEDCS
Sbjct: 397 LDLLFHGAQVHRHAHNFREVQQCTLLSIKTGGCSEDCS 510
>AW309471
Length = 400
Score = 137 bits (346), Expect = 6e-33
Identities = 76/126 (60%), Positives = 83/126 (65%)
Frame = -1
Query: 239 EAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPK 298
+ E R H T ESV IN L+ VKG +D K VEIWEMIRMIAT RI M K
Sbjct: 379 QEEQPRAAPSHPSPTFLPTSESVLINVLVXVKGIFFEDXKFVEIWEMIRMIATVRIIMLK 200
Query: 299 AMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLPKAPS 358
MVRL AGRVRF + E LCFLAG N IF GEK+ TT NNDFDAD LMFKV GL PKAP
Sbjct: 199 VMVRLLAGRVRFFMLE*VLCFLAGVNFIFTGEKVFTTLNNDFDAD*LMFKVFGLFPKAPR 20
Query: 359 LDDDET 364
L++ T
Sbjct: 19 LNEGAT 2
>BU965020
Length = 416
Score = 80.9 bits (198), Expect = 9e-16
Identities = 40/48 (83%), Positives = 43/48 (89%)
Frame = +2
Query: 330 EKLLTTANNDFDADQLMFKVLGLLPKAPSLDDDETNEAENYKEAASSS 377
EKLLTT NNDFDADQLMFKVLGLLPKAPSL++ ETN E+YKEAA SS
Sbjct: 2 EKLLTTPNNDFDADQLMFKVLGLLPKAPSLNEGETNVTEDYKEAAFSS 145
>TC203398 weakly similar to UP|Q7XYK7 (Q7XYK7) Ribosomal protein rpL31,
partial (28%)
Length = 879
Score = 31.2 bits (69), Expect = 0.80
Identities = 13/41 (31%), Positives = 20/41 (48%), Gaps = 2/41 (4%)
Frame = -2
Query: 71 HRHAHNFREVQQCTLLSVK--TGGCSEDCSYCPQSSRYDTG 109
H+ H R + C + ++K T GC +CP + DTG
Sbjct: 743 HQTRHGHRHIDTCNVQNIKHNTDGCRVGVGHCPTPTMSDTG 621
>TC224929 homologue to UP|Q6RYC4 (Q6RYC4) 60S ribosomal protein L19, partial
(80%)
Length = 917
Score = 30.4 bits (67), Expect = 1.4
Identities = 20/58 (34%), Positives = 27/58 (46%)
Frame = +1
Query: 35 AEKTIQNGPRNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGG 92
AEK G R+DW K K P+L LL H +Q +H N R +L+ + G
Sbjct: 526 AEKEKLLGGRSDWPKVLEKKYPLHPVLQLLQHHSQ-RKHQRNLRSDPHILILACRIFG 696
>BF324334
Length = 417
Score = 30.0 bits (66), Expect = 1.8
Identities = 21/61 (34%), Positives = 30/61 (48%), Gaps = 7/61 (11%)
Frame = +1
Query: 221 FVRDAGINVCSGGIIGLGEA-------EDDRVGLLHTLSTLPTHPESVPINALIAVKGTP 273
FVR N+CSGG++ GE E DR+ L +P P S + ++A +G P
Sbjct: 37 FVRFTISNICSGGLLNAGETEALEETPEPDRISL-----RIPLIPRS---SKVLAPRGDP 192
Query: 274 L 274
L
Sbjct: 193 L 195
>TC228496
Length = 762
Score = 28.9 bits (63), Expect = 4.0
Identities = 13/33 (39%), Positives = 18/33 (54%)
Frame = +3
Query: 81 QQCTLLSVKTGGCSEDCSYCPQSSRYDTGLKGQ 113
++ TL+ GGC E CS+CP + D GQ
Sbjct: 177 EEITLVQKFRGGCEEACSHCPFFAFGDKQCLGQ 275
>TC230108 similar to UP|CNX2_ARATH (Q39055) Molybdopterin biosynthesis CNX2
protein (Molybdenum cofactor biosynthesis enzyme CNX2),
partial (49%)
Length = 849
Score = 28.1 bits (61), Expect = 6.8
Identities = 35/157 (22%), Positives = 61/157 (38%), Gaps = 1/157 (0%)
Frame = +1
Query: 90 TGGCSEDCSYCPQSSRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRK 149
T C+ C YC + + Q LL K +L+ A +G + + TI +
Sbjct: 358 TERCNLRCQYCMPADGVELTPSPQ-LLTKTEILRCANLFVSSGVNKIRLTGG-EPTI--R 525
Query: 150 TNFNQILEYVKEIKGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITT 209
+ I + +KG+ T G+ + +LK+ GL + N +LDT +T
Sbjct: 526 KDIEDICLELSTLKGLKTLSMTTNGIALARKLPKLKECGLNSVNISLDTLVPAKFEFMTR 705
Query: 210 RTYDER-LKTLEFVRDAGINVCSGGIIGLGEAEDDRV 245
R E+ + + D G N + + DD +
Sbjct: 706 RKGHEKVMDAINASIDLGFNPVKVNCVVMRGFNDDEI 816
>TC203926 weakly similar to PIR|T06696|T06696 lipase homolog T29H11.20 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(54%)
Length = 855
Score = 27.7 bits (60), Expect = 8.9
Identities = 24/88 (27%), Positives = 37/88 (41%), Gaps = 17/88 (19%)
Frame = +2
Query: 162 IKGMGMEVCCTLGML-----DKDQAGELKKAGLTAYNHN------LDTSREYYPN-IITT 209
++G+ + C TL M D+D G +K +Y HN L R+ YP +I
Sbjct: 194 VQGLPLTGCLTLSMYLAPPDDRDDIGCVKSVNNQSYYHNLVLQDKLQEFRKQYPQAVILY 373
Query: 210 RTYDERLKTL-----EFVRDAGINVCSG 232
Y + +T+ +F NVC G
Sbjct: 374 ADYYDAYRTVMKNPSKFGFKETFNVCCG 457
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,802,565
Number of Sequences: 63676
Number of extensions: 219672
Number of successful extensions: 1122
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 1118
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1122
length of query: 377
length of database: 12,639,632
effective HSP length: 99
effective length of query: 278
effective length of database: 6,335,708
effective search space: 1761326824
effective search space used: 1761326824
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146331.7


