

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145767.3 - phase: 0
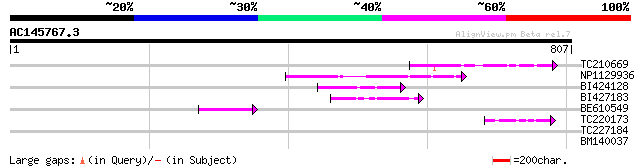
(807 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC210669 similar to UP|Q9FFR9 (Q9FFR9) Na+/H+ antiporter-like pr... 118 1e-26
NP1129936 CHX3 [Glycine max] 91 3e-18
BI424128 similar to GP|10178015|db Na+/H+ antiporter-like protei... 67 2e-11
BI427183 61 2e-09
BE610549 58 2e-08
TC220173 48 2e-05
TC227184 UP|Q6Q2Z9 (Q6Q2Z9) Phosphoenolpyruvate carboxylase , p... 31 1.9
BM140037 29 9.4
>TC210669 similar to UP|Q9FFR9 (Q9FFR9) Na+/H+ antiporter-like protein,
partial (16%)
Length = 952
Score = 118 bits (295), Expect = 1e-26
Identities = 83/219 (37%), Positives = 119/219 (53%), Gaps = 6/219 (2%)
Frame = +3
Query: 576 GSPEISHNEHCEINENVLQHAPCSVGIFVDRGLG-----SLLKTKMRIITLFIGGPDDRE 630
GS I+ N+ +N+ VL+HAPCSVGIFVDRGLG S R+ LF GG DDRE
Sbjct: 15 GSLNITRNDXRWVNKRVLEHAPCSVGIFVDRGLGGTSHVSASNVSYRVTVLFFGGGDDRE 194
Query: 631 ALSIAWRMAGHSGTQLHVVRIHLLGKAAEE-KVLKKKISKSPHGMLSTVMDGVMQKELDE 689
AL+ RMA H G +L V+R +G+ E ++++ + S G DE
Sbjct: 195 ALAYGARMAEHPGIRLLVIR--FVGEPMNEGEIVRVDVGDS---------TGTKLISQDE 341
Query: 690 EYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIF 749
E++ F+ K + N+DSI+Y EK V G E ++ E++ +L++VG
Sbjct: 342 EFLDEFKAK-IANDDSIIYEEKVVKD--GAETVAIICELN--SCNLFLVGSRPASEVASA 506
Query: 750 LKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQQYMVG 788
+K E PELG +G +LAS + T +SVL++QQY G
Sbjct: 507 MKRSEC---PELGPVGGLLASQDYPTTASVLVMQQYQNG 614
>NP1129936 CHX3 [Glycine max]
Length = 1125
Score = 90.5 bits (223), Expect = 3e-18
Identities = 74/260 (28%), Positives = 124/260 (47%), Gaps = 1/260 (0%)
Frame = +1
Query: 398 SFMVMTLAIILMTVIVSPLINAIYKPKFRFMQSQLRTVQKLRFDMELRVVACVHNAKHAT 457
++ VM + I+++ IV + IY P ++ R + L+ D ELRVVAC+H HA+
Sbjct: 313 TYGVMMINIMVIASIVKWSVKLIYDPSRKYAGYPKRNIASLKPDSELRVVACLHKTHHAS 492
Query: 458 NIIHVLEATNATRISPVHVSAVHLLELTRHGTAILVSQMADLNNIAGAETTNYGPKAEFE 517
+ L+ T P + S+ H K+ +
Sbjct: 493 AVKDCLDLCCPTTEDP-NCSSNH--------------------------------KSYSD 573
Query: 518 IITTAFKEFV-EQYNAVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGG 576
I AF + + AV + +S + +HED+ ++A +K AS+I+LPFH +S G
Sbjct: 574 DIILAFDLYEHDNMGAVTAHVYTAISPPSLMHEDVCHLALDKVASIIILPFHLRWSG-DG 750
Query: 577 SPEISHNEHCEINENVLQHAPCSVGIFVDRGLGSLLKTKMRIITLFIGGPDDREALSIAW 636
+ E + +N +L+ APCSVGI V R + +++ +F+GG DDREAL +A
Sbjct: 751 AIESDYKNARALNCKLLEIAPCSVGILVGRS-AIHCDSFIQVAMIFLGGNDDREALCLAK 927
Query: 637 RMAGHSGTQLHVVRIHLLGK 656
R+ ++++V HL+ K
Sbjct: 928 RVT--RNPRVNLVVYHLVPK 981
>BI424128 similar to GP|10178015|db Na+/H+ antiporter-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 421
Score = 67.4 bits (163), Expect = 2e-11
Identities = 43/128 (33%), Positives = 68/128 (52%), Gaps = 1/128 (0%)
Frame = +1
Query: 443 ELRVVACVHNAKHATNIIHVLEATNATRISP-VHVSAVHLLELTRHGTAILVSQMADLNN 501
+LR++ C H A++ ++I+++EA+ R + V A+HL E + + IL+ A N
Sbjct: 25 QLRILTCFHGARNIPSMINLIEASRGIRKGDALCVYAMHLKEFSERSSTILMVHKARRN- 201
Query: 502 IAGAETTNYGPKAEFEIITTAFKEFVEQYNAVRFDTSSVVSSYTTIHEDIYNVAEEKRAS 561
G N G A+ + AF+ + Q + V +SS IHEDI AE K A+
Sbjct: 202 --GLPFWNKGHHADSNHVIVAFEAY-RQLSQVSIRPMIAISSMNNIHEDICATAERKGAA 372
Query: 562 LILLPFHK 569
+I+LPFHK
Sbjct: 373 VIILPFHK 396
>BI427183
Length = 421
Score = 61.2 bits (147), Expect = 2e-09
Identities = 40/134 (29%), Positives = 72/134 (52%)
Frame = +2
Query: 462 VLEATNATRISPVHVSAVHLLELTRHGTAILVSQMADLNNIAGAETTNYGPKAEFEIITT 521
++E+ + + S + + +HL+ELT ++I+++Q D N +G+ + E +
Sbjct: 2 LIESIRSIQKSSIKLFIMHLVELTERSSSIILAQNTD--NKSGSSHVEW-----LEQLYR 160
Query: 522 AFKEFVEQYNAVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEIS 581
AF+ Q V + + +SS +T+H+DI +VA+EK ++I+LPFHK + V
Sbjct: 161 AFQAH-SQLGQVSVQSKTTISSLSTMHDDICHVADEKMVTMIILPFHKRWKKV------- 316
Query: 582 HNEHCEINENVLQH 595
E+ E N V QH
Sbjct: 317 EMENEEENSEVSQH 358
>BE610549
Length = 385
Score = 57.8 bits (138), Expect = 2e-08
Identities = 29/86 (33%), Positives = 49/86 (56%), Gaps = 1/86 (1%)
Frame = -2
Query: 272 ITDFLGTHPIVGAFVFGLILPHG-KFADMVMEMSDDFVTGILCPVYFSGFGFKLDLPVLW 330
+T +G H I G FVFGL +P G +FA+ + +DFV+ + P+YF+ G K D+ L
Sbjct: 372 VTHMIGLHSIFGGFVFGLTIPKGGEFANRMTRRIEDFVSTLFLPLYFAASGLKTDVTKLR 193
Query: 331 NTPNSVLMMLIMVLLCIPKVLSSLIV 356
+ + L++L+ + K+L + V
Sbjct: 192 SVVDWGLLLLVTSTASVGKILGTFAV 115
>TC220173
Length = 567
Score = 48.1 bits (113), Expect = 2e-05
Identities = 38/102 (37%), Positives = 59/102 (57%)
Frame = +1
Query: 684 QKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIPTLLNEIDKPGYDLYIVGQGSG 743
+KELD+ + F+ K + +V ++V S+ EE+ L D YDL IVG+G
Sbjct: 127 EKELDDATMARFQRKW----NGMVECFEKVASNIMEEVLALGRSKD---YDLIIVGKGQF 285
Query: 744 KNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQQY 785
+ ++ L++ H ELG IGDILAS++ SSVL++QQ+
Sbjct: 286 -SLSLVADLVDR-QHEELGPIGDILASSTHDVVSSVLVIQQH 405
>TC227184 UP|Q6Q2Z9 (Q6Q2Z9) Phosphoenolpyruvate carboxylase , partial (63%)
Length = 2132
Score = 31.2 bits (69), Expect = 1.9
Identities = 28/123 (22%), Positives = 49/123 (39%), Gaps = 9/123 (7%)
Frame = +1
Query: 662 VLKKKISKSPHGMLSTVMDGVMQ-------KELDEEYIFSFRHKAVNNNDSIVYLEKEVH 714
VL K + G + +D + Q E DEE F + + +V + EVH
Sbjct: 427 VLMKLDLRQESGRHAEALDAITQYLDMGTYSEWDEEKKLDFLTRELKGKRPLVPVSIEVH 606
Query: 715 SDTGEEIPT--LLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTS 772
D E + T + E+ Y++ S + + ++LL+ L IG++ +
Sbjct: 607 PDVKEVLDTFRIAAELGSDSLGAYVISMASNASDVLAVELLQ--KDARLAAIGELGKACP 780
Query: 773 FGT 775
GT
Sbjct: 781 GGT 789
>BM140037
Length = 272
Score = 28.9 bits (63), Expect = 9.4
Identities = 19/52 (36%), Positives = 32/52 (61%), Gaps = 1/52 (1%)
Frame = -3
Query: 733 YDLYIVGQGSGKNKTIFLKLLEWCD-HPELGVIGDILASTSFGTHSSVLIVQ 783
+DL IVG+ + +T L +++ + ELG IGD+ S+ G SS+L++Q
Sbjct: 210 HDLVIVGKQ--QLETTMLTNIDFRHGNEELGPIGDLFVSSGNGITSSLLVIQ 61
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.140 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,562,304
Number of Sequences: 63676
Number of extensions: 506724
Number of successful extensions: 3307
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 3276
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3300
length of query: 807
length of database: 12,639,632
effective HSP length: 105
effective length of query: 702
effective length of database: 5,953,652
effective search space: 4179463704
effective search space used: 4179463704
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC145767.3


