

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145767.1 - phase: 0
(191 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
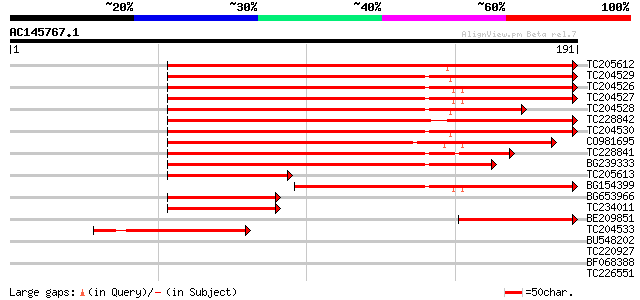
Score E
Sequences producing significant alignments: (bits) Value
TC205612 similar to GB|AAM91395.1|22137100|AY133565 At4g14410/dl... 249 5e-67
TC204529 150 3e-37
TC204526 146 5e-36
TC204527 145 8e-36
TC204528 127 2e-30
TC228842 124 3e-29
TC204530 121 2e-28
CO981695 109 7e-25
TC228841 100 3e-22
BG239333 100 7e-22
TC205613 82 1e-16
BG154399 82 2e-16
BG653966 52 1e-07
TC234011 48 2e-06
BE209851 47 7e-06
TC204533 weakly similar to UP|SFRB_HUMAN (Q05519) Splicing facto... 44 6e-05
BU548202 35 0.016
TC220927 similar to UP|Q9LT23 (Q9LT23) Emb|CAA18500.1, partial (... 33 0.10
BF068388 weakly similar to GP|29367409|gb| helix-loop-helix-like... 33 0.10
TC226551 similar to GB|AAP37829.1|30725614|BT008470 At5g12470 {A... 32 0.14
>TC205612 similar to GB|AAM91395.1|22137100|AY133565 At4g14410/dl3245w
{Arabidopsis thaliana;} , partial (47%)
Length = 863
Score = 249 bits (636), Expect = 5e-67
Identities = 124/141 (87%), Positives = 131/141 (91%), Gaps = 3/141 (2%)
Frame = +3
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RFCDLS+VLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELK++NEKLLEEIKCLKAEKNE
Sbjct: 216 RFCDLSSVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKKTNEKLLEEIKCLKAEKNE 395
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPP---PMAAYQASVNKMAVYPNYGYIPMW 170
LREEKLVLKADKE+IEKQLK++PV+PAGFM PP AAYQA VNKMAVYPNYGYIPMW
Sbjct: 396 LREEKLVLKADKERIEKQLKALPVAPAGFMAPPVAAAAAAYQAGVNKMAVYPNYGYIPMW 575
Query: 171 HYLPQSARDTSQDHELRPPAA 191
YLPQS RDTS DHELRPPAA
Sbjct: 576 QYLPQSVRDTSHDHELRPPAA 638
>TC204529
Length = 854
Score = 150 bits (380), Expect = 3e-37
Identities = 79/144 (54%), Positives = 106/144 (72%), Gaps = 6/144 (4%)
Frame = +1
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF +L ++LEPGRP +TDK AIL DA+R+++QL+ EAQ+LK++N+ L E+IK LKAEKNE
Sbjct: 70 RFVELGSILEPGRPAKTDKTAILIDAVRMVTQLRGEAQKLKDTNQGLQEKIKELKAEKNE 249
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPP------MAAYQASVNKMAVYPNYGYI 167
LR+EK LKA+KEK+E+QLKS+ P+ FMPPP A QA NK+ + +Y +
Sbjct: 250 LRDEKQRLKAEKEKLEQQLKSLNAQPS-FMPPPAAIPAAFAAQGQAHGNKLVPFISYPGV 426
Query: 168 PMWHYLPQSARDTSQDHELRPPAA 191
MW ++P +A DTSQDH LRPP A
Sbjct: 427 AMWQFMPPAAVDTSQDHVLRPPVA 498
>TC204526
Length = 807
Score = 146 bits (369), Expect = 5e-36
Identities = 79/144 (54%), Positives = 105/144 (72%), Gaps = 6/144 (4%)
Frame = +2
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L ++LEPGRP +TDK +IL DA R+++QL+ EA +LK+SN L E+IK LKAEKNE
Sbjct: 83 KFVELGSILEPGRPPKTDKASILIDAARMVTQLRDEALKLKDSNTSLQEKIKELKAEKNE 262
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPM--AAY----QASVNKMAVYPNYGYI 167
LR+EK LKA+KEK+E Q+KSM PA F+PPPP AA+ QA NK+ + +Y +
Sbjct: 263 LRDEKQRLKAEKEKLEVQVKSMNAQPA-FLPPPPAIPAAFAPQGQAPGNKLVPFISYPGV 439
Query: 168 PMWHYLPQSARDTSQDHELRPPAA 191
MW ++P +A DTSQDH LRPP A
Sbjct: 440 AMWQFMPPAAVDTSQDHVLRPPVA 511
>TC204527
Length = 898
Score = 145 bits (367), Expect = 8e-36
Identities = 79/144 (54%), Positives = 105/144 (72%), Gaps = 6/144 (4%)
Frame = +2
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L ++LEPGRP +TDK +IL DA R+++QL+ EA +LK+SN L E+IK LKAEKNE
Sbjct: 470 KFVELGSILEPGRPPKTDKASILIDAARMVTQLRDEALKLKDSNTSLQEKIKELKAEKNE 649
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPM--AAY----QASVNKMAVYPNYGYI 167
LR+EK LKA+KEK+E Q+KSM PA F+PPPP AA+ QA NK+ + +Y +
Sbjct: 650 LRDEKQRLKAEKEKLEVQVKSMNAQPA-FLPPPPAIPAAFAPQGQAPGNKLVPFISYPGV 826
Query: 168 PMWHYLPQSARDTSQDHELRPPAA 191
MW ++P +A DTSQDH LRPP A
Sbjct: 827 AMWQFMPPAAVDTSQDHVLRPPDA 898
>TC204528
Length = 793
Score = 127 bits (320), Expect = 2e-30
Identities = 67/127 (52%), Positives = 93/127 (72%), Gaps = 6/127 (4%)
Frame = +2
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L A+LEPGRP +TDK AIL DA+R+++QL+ EAQ+LK++N+ L E+IK LKAEKNE
Sbjct: 410 KFVELGAILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDTNQGLQEKIKELKAEKNE 589
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPP------MAAYQASVNKMAVYPNYGYI 167
LR+EK LKA+KEK+E+QLKS+ P+ FMPPP A QA NK+ + +Y +
Sbjct: 590 LRDEKQRLKAEKEKLEQQLKSLNAQPS-FMPPPAAMPAAFAAQGQAHGNKLVPFISYPGV 766
Query: 168 PMWHYLP 174
MW ++P
Sbjct: 767 AMWQFMP 787
>TC228842
Length = 1023
Score = 124 bits (310), Expect = 3e-29
Identities = 64/138 (46%), Positives = 92/138 (66%)
Frame = +2
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF +L ++L+P +P++ DK IL DA+RV+SQL+ EAQ+L+ES E L E+I LK EKNE
Sbjct: 365 RFMELGSILDPRKPLKMDKAVILSDAVRVVSQLREEAQKLRESTENLQEKINALKDEKNE 544
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPMAAYQASVNKMAVYPNYGYIPMWHYL 173
LR+EK LK +KE +E+++K++ P+ AA Q +K+ + Y + MW +L
Sbjct: 545 LRDEKQRLKVEKENLEQKVKALSSQPSFL-----AAAGQVVGSKLVPFMGYPGVAMWQFL 709
Query: 174 PQSARDTSQDHELRPPAA 191
+A DTSQDH LRPP A
Sbjct: 710 SPAAVDTSQDHVLRPPVA 763
>TC204530
Length = 957
Score = 121 bits (303), Expect = 2e-28
Identities = 68/144 (47%), Positives = 94/144 (65%), Gaps = 6/144 (4%)
Frame = +1
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L ++LEPGRP +TDK AIL DA ++ +Q + E+K++N+ L E+IK LKAEKNE
Sbjct: 529 KFVELGSILEPGRPAKTDKAAILIDAGQMGNQFTG*SPEVKDTNQGLQEKIKELKAEKNE 708
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPP------MAAYQASVNKMAVYPNYGYI 167
LR+E+ LKA+ EK+E+QL+S+ P+ FMPP A QA NK+ + +
Sbjct: 709 LRDERHRLKAENEKLEQQLRSVNAQPS-FMPPAAAMPAACAAQGQAHGNKVVPFISNPRS 885
Query: 168 PMWHYLPQSARDTSQDHELRPPAA 191
MW Y+P RDTSQDH L PP A
Sbjct: 886 AMWQYMPPGCRDTSQDHILHPPVA 957
>CO981695
Length = 415
Score = 109 bits (273), Expect = 7e-25
Identities = 62/137 (45%), Positives = 91/137 (66%), Gaps = 6/137 (4%)
Frame = -1
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L ++ E GR +TDK IL D +R++SQL+ EAQ+LK+ N+ L E+I+ LK EKNE
Sbjct: 412 KFVELGSI*ERGRHAKTDKSXILIDDVRMVSQLRGEAQKLKDXNQGLQEKIRDLKXEKNE 233
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPP--PPMAAY----QASVNKMAVYPNYGYI 167
+R++K +KA+KEK+E+QLKS+ FMPP AA+ QA NK+ + +
Sbjct: 232 VRDQKQRIKAEKEKLEQQLKSLN-GQLSFMPPRDAXXAAFAVQGQAHGNKLVPFIRDTGV 56
Query: 168 PMWHYLPQSARDTSQDH 184
MW ++P +A DTSQDH
Sbjct: 55 AMWQFMPLAAVDTSQDH 5
>TC228841
Length = 584
Score = 100 bits (250), Expect = 3e-22
Identities = 55/117 (47%), Positives = 81/117 (69%)
Frame = +3
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF +L +++ PG P++ DK IL DA+RV+SQL+ EAQ+L+ES+E L E+I LKAEKNE
Sbjct: 240 RFMELWSIVGPGMPLKMDKAVILSDAVRVVSQLQEEAQKLRESSENLQEKINELKAEKNE 419
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPMAAYQASVNKMAVYPNYGYIPMW 170
LR+EK LKA+K+ IE++L ++ P+ F+P P A Q +K+ + Y + MW
Sbjct: 420 LRDEKQRLKAEKDSIEQKLIALSSQPS-FLPAFPSAG-QVVGSKLVPFMGYPGVAMW 584
>BG239333
Length = 376
Score = 99.8 bits (247), Expect = 7e-22
Identities = 53/111 (47%), Positives = 76/111 (67%)
Frame = +2
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF +LS++LEP R ++DK AIL DA RV+ QL+ EA+ LKE N++L ++K LK EKNE
Sbjct: 41 RFLELSSILEPSRQPKSDKVAILSDAARVVIQLRNEAKRLKEMNDELQAKVKELKGEKNE 220
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPMAAYQASVNKMAVYPNY 164
LR+EK LK +KEK+E+Q+K + P+ F+P P A Q +K+ + Y
Sbjct: 221 LRDEKNRLKEEKEKLEQQVKVANIQPS-FLPQAPDAKGQVGSHKLIPFIGY 370
>TC205613
Length = 918
Score = 82.4 bits (202), Expect = 1e-16
Identities = 40/42 (95%), Positives = 42/42 (99%)
Frame = +3
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKE 95
RFCDLS+VLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELK+
Sbjct: 792 RFCDLSSVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKK 917
>BG154399
Length = 301
Score = 82.0 bits (201), Expect = 2e-16
Identities = 47/101 (46%), Positives = 64/101 (62%), Gaps = 6/101 (5%)
Frame = +2
Query: 97 NEKLLEEIKCLKAEKNELREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPM--AAY--- 151
N L E++K LKAEKNEL ++K +LKA K+K+E Q+KSM P F+PPP AA+
Sbjct: 2 NTSLKEKMKDLKAEKNELSDQKQMLKA*KDKLEVQVKSMNAKPV-FLPPPSAIPAAFAPQ 178
Query: 152 -QASVNKMAVYPNYGYIPMWHYLPQSARDTSQDHELRPPAA 191
Q+ NK+ + +Y + MW ++P DT QDH LRPP A
Sbjct: 179 GQSDGNKLVPFISYPGVAMWLFMPADVVDT*QDHVLRPPVA 301
>BG653966
Length = 416
Score = 52.4 bits (124), Expect = 1e-07
Identities = 23/38 (60%), Positives = 32/38 (83%)
Frame = +1
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQ 91
RF +L ++LEPGRP +TDK AIL DA+R+++QL+ EAQ
Sbjct: 301 RFVELGSILEPGRPAKTDKTAILIDAVRMVTQLRGEAQ 414
>TC234011
Length = 473
Score = 48.1 bits (113), Expect = 2e-06
Identities = 22/38 (57%), Positives = 30/38 (78%)
Frame = +2
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQ 91
RF +LS++LEPGR +TDK A+L DA RV+ QL+ EA+
Sbjct: 356 RFLELSSILEPGRQPKTDKVALLSDAARVVIQLRNEAE 469
>BE209851
Length = 379
Score = 46.6 bits (109), Expect = 7e-06
Identities = 20/40 (50%), Positives = 27/40 (67%)
Frame = +1
Query: 152 QASVNKMAVYPNYGYIPMWHYLPQSARDTSQDHELRPPAA 191
QA NK+ + +Y + MW ++P +A DTSQDH LRPP A
Sbjct: 121 QAPGNKLVPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 240
>TC204533 weakly similar to UP|SFRB_HUMAN (Q05519) Splicing factor
arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear
protein) (p54), partial (5%)
Length = 663
Score = 43.5 bits (101), Expect = 6e-05
Identities = 26/53 (49%), Positives = 34/53 (64%)
Frame = +1
Query: 29 LLSVLYCVLLLHMIKGCSFIVIVCCRFCDLSAVLEPGRPVRTDKPAILDDAIR 81
LLS+ CV +M G + +I RF +L ++LEPGRP +TDK AIL DA R
Sbjct: 514 LLSLWVCV---NMFWGHASNMIDLHRFVELGSILEPGRPPKTDKAAILIDAAR 663
>BU548202
Length = 576
Score = 35.4 bits (80), Expect = 0.016
Identities = 19/52 (36%), Positives = 33/52 (62%)
Frame = -3
Query: 83 LSQLKTEAQELKESNEKLLEEIKCLKAEKNELREEKLVLKADKEKIEKQLKS 134
L LK E +LKE +EK +EIK LK E + L + +K++ E+ +K++++
Sbjct: 505 LENLKKETVQLKEKDEKASKEIKQLKEELSCLSKSLEKIKSESEEKDKKVET 350
>TC220927 similar to UP|Q9LT23 (Q9LT23) Emb|CAA18500.1, partial (12%)
Length = 700
Score = 32.7 bits (73), Expect = 0.10
Identities = 17/30 (56%), Positives = 20/30 (66%)
Frame = +1
Query: 102 EEIKCLKAEKNELREEKLVLKADKEKIEKQ 131
EE + L EKNELREEK LK+D E + Q
Sbjct: 1 EESRELMQEKNELREEKTSLKSDIENLNVQ 90
>BF068388 weakly similar to GP|29367409|gb| helix-loop-helix-like protein
{Oryza sativa (japonica cultivar-group)}, partial (12%)
Length = 424
Score = 32.7 bits (73), Expect = 0.10
Identities = 12/27 (44%), Positives = 20/27 (73%)
Frame = +1
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAI 80
RF +L ++L+P +P++ DK IL DA+
Sbjct: 343 RFMELGSILDPRKPLKMDKAVILSDAV 423
>TC226551 similar to GB|AAP37829.1|30725614|BT008470 At5g12470 {Arabidopsis
thaliana;} , partial (75%)
Length = 1883
Score = 32.3 bits (72), Expect = 0.14
Identities = 18/51 (35%), Positives = 30/51 (58%), Gaps = 6/51 (11%)
Frame = +2
Query: 108 KAEKNELREEKLVLKADKEKIEKQLKSM-----PVSP-AGFMPPPPMAAYQ 152
K +K + ++EK K K+K +K+ K+ P++P GF PPPP A ++
Sbjct: 1574 KKKKKKKKKEKKKKKKKKKKKKKKKKTRGGARAPIAPFGGFFPPPPGAVFK 1726
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,438,258
Number of Sequences: 63676
Number of extensions: 143153
Number of successful extensions: 1324
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 1293
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1304
length of query: 191
length of database: 12,639,632
effective HSP length: 92
effective length of query: 99
effective length of database: 6,781,440
effective search space: 671362560
effective search space used: 671362560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC145767.1


