

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145512.8 - phase: 0 /pseudo
(164 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
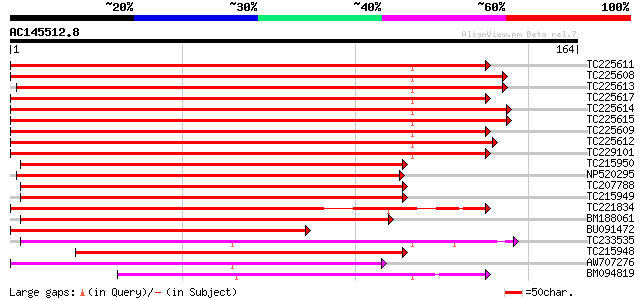
Score E
Sequences producing significant alignments: (bits) Value
TC225611 homologue to UP|Q84ZU6 (Q84ZU6) R 1 protein, complete 144 1e-35
TC225608 UP|Q84ZU5 (Q84ZU5) R 8 protein, complete 142 5e-35
TC225613 UP|Q9FVK5 (Q9FVK5) Resistance protein LM6 (Fragment), p... 142 9e-35
TC225617 UP|Q84ZV8 (Q84ZV8) R 3 protein, complete 140 4e-34
TC225614 UP|Q84ZV7 (Q84ZV7) R 12 protein, partial (88%) 139 8e-34
TC225615 UP|Q84ZU7 (Q84ZU7) R 5 protein, complete 138 1e-33
TC225609 UP|Q84ZV3 (Q84ZV3) R 4 protein, complete 137 2e-33
TC225612 homologue to UP|Q84ZU8 (Q84ZU8) R 10 protein, complete 137 2e-33
TC229101 disease resistance-like protein 129 8e-31
TC215950 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candida... 125 9e-30
NP520295 disease resistance-like protein GS5-3 [Glycine max] 125 1e-29
TC207788 homologue to UP|Q8W2C0 (Q8W2C0) Functional candidate re... 114 2e-26
TC215949 similar to UP|Q8W2C0 (Q8W2C0) Functional candidate resi... 113 5e-26
TC221834 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candida... 111 1e-25
BM188061 weakly similar to GP|18033111|g functional candidate re... 110 2e-25
BU091472 similar to GP|9965103|gb|A resistance protein LM6 {Glyc... 107 2e-24
TC233535 weakly similar to UP|Q6XZH4 (Q6XZH4) Nematode resistanc... 101 2e-22
TC215948 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candida... 95 2e-20
AW707276 similar to GP|9858483|gb|A resistance protein MG55 {Gly... 72 9e-14
BM094819 weakly similar to PIR|T47438|T47 disease resistance pro... 69 1e-12
>TC225611 homologue to UP|Q84ZU6 (Q84ZU6) R 1 protein, complete
Length = 2709
Score = 144 bits (364), Expect = 1e-35
Identities = 81/140 (57%), Positives = 93/140 (65%), Gaps = 1/140 (0%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
+ VLNFD LTQIPDVS L NL+E SF C +LI V DSIG L +LK L C KLR
Sbjct: 1888 LTVLNFDQCEFLTQIPDVSDLPNLKELSFDWCESLIAVDDSIGFLNKLKKLSAYGCRKLR 2067
Query: 61 SIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKI 119
S PPLNL SLE L LS CS LE FPEILGEM N+ L+L+ PIKELPF FQNL L ++
Sbjct: 2068 SFPPLNLTSLETLQLSGCSSLEYFPEILGEMENIKALDLDGLPIKELPFSFQNLIGLCRL 2247
Query: 120 YPCNCGFVSLPTSNYAMSKL 139
+CG + LP S M +L
Sbjct: 2248 TLNSCGIIQLPCSLAMMPEL 2307
>TC225608 UP|Q84ZU5 (Q84ZU5) R 8 protein, complete
Length = 3084
Score = 142 bits (359), Expect = 5e-35
Identities = 82/145 (56%), Positives = 94/145 (64%), Gaps = 1/145 (0%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
+ VLNFD LT+IPDVS L NL+E SF C +L+ V DSIG L +LKTL C KL
Sbjct: 1885 LTVLNFDRCEFLTKIPDVSDLPNLKELSFNWCESLVAVDDSIGFLNKLKTLSAYGCRKLT 2064
Query: 61 SIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKI 119
S PPLNL SLE L+L CS LE FPEILGEM+N+T L L PIKELPF FQNL L +
Sbjct: 2065 SFPPLNLTSLETLNLGGCSSLEYFPEILGEMKNITVLALHDLPIKELPFSFQNLIGLLFL 2244
Query: 120 YPCNCGFVSLPTSNYAMSKLVECTI 144
+ +CG V L S M KL E I
Sbjct: 2245 WLDSCGIVQLRCSLATMPKLCEFCI 2319
>TC225613 UP|Q9FVK5 (Q9FVK5) Resistance protein LM6 (Fragment), partial (98%)
Length = 2597
Score = 142 bits (357), Expect = 9e-35
Identities = 81/143 (56%), Positives = 93/143 (64%), Gaps = 1/143 (0%)
Frame = +1
Query: 3 VLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSI 62
+LNFD LT+IPDVS L NL+E SF C +L+ V DSIG L +LKTL C KL S
Sbjct: 1852 ILNFDRCEFLTKIPDVSDLPNLKELSFNWCESLVAVDDSIGFLNKLKTLSAYGCRKLTSF 2031
Query: 63 PPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKIYP 121
PPLNL SLE L+L CS LE FPEILGEM+N+T L L PIKELPF FQNL L ++
Sbjct: 2032 PPLNLTSLETLNLGGCSSLEYFPEILGEMKNITVLALHDLPIKELPFSFQNLIGLLFLWL 2211
Query: 122 CNCGFVSLPTSNYAMSKLVECTI 144
+CG V L S M KL E I
Sbjct: 2212 DSCGIVQLRCSLATMPKLCEFCI 2280
>TC225617 UP|Q84ZV8 (Q84ZV8) R 3 protein, complete
Length = 2697
Score = 140 bits (352), Expect = 4e-34
Identities = 80/140 (57%), Positives = 89/140 (63%), Gaps = 1/140 (0%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
+ VL FD LTQIPDVS L NL E SFQ C +L+ V DSIG L +LK L C KL
Sbjct: 1882 LTVLKFDWCKFLTQIPDVSDLPNLRELSFQWCESLVAVDDSIGFLKKLKKLNADGCRKLT 2061
Query: 61 SIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKI 119
S PPLNL SLE L+LS CS LE FPEILGEM N+ L+L PIKELPF FQNL L ++
Sbjct: 2062 SFPPLNLTSLETLELSHCSSLEYFPEILGEMENIERLDLHGLPIKELPFSFQNLIGLQQL 2241
Query: 120 YPCNCGFVSLPTSNYAMSKL 139
CG V L S M KL
Sbjct: 2242 SMFGCGIVQLRCSLAMMPKL 2301
>TC225614 UP|Q84ZV7 (Q84ZV7) R 12 protein, partial (88%)
Length = 2367
Score = 139 bits (349), Expect = 8e-34
Identities = 80/146 (54%), Positives = 91/146 (61%), Gaps = 1/146 (0%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
+ VL FD LTQIPDVS L NL E SF GC +L+ + DSIG L +L+ L C KL
Sbjct: 1870 LTVLKFDKCKFLTQIPDVSDLPNLRELSFVGCESLVAIDDSIGFLNKLEILNAAGCRKLT 2049
Query: 61 SIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKI 119
S PPLNL SLE L+LS CS LE FPEILGEM N+T L+LE PIKELPF FQNL L +I
Sbjct: 2050 SFPPLNLTSLETLELSHCSSLEYFPEILGEMENITALHLERLPIKELPFSFQNLIGLREI 2229
Query: 120 YPCNCGFVSLPTSNYAMSKLVECTIQ 145
C V L S M L I+
Sbjct: 2230 TLRRCRIVRLRCSLAMMPNLFRFQIR 2307
>TC225615 UP|Q84ZU7 (Q84ZU7) R 5 protein, complete
Length = 2724
Score = 138 bits (347), Expect = 1e-33
Identities = 79/146 (54%), Positives = 91/146 (62%), Gaps = 1/146 (0%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
+ VL FD+ LTQIPDVS L NL E SF+GC +L+ V DSIG L +LK L C KL
Sbjct: 1876 LTVLKFDNCKFLTQIPDVSDLPNLRELSFKGCESLVAVDDSIGFLNKLKKLNAYGCRKLT 2055
Query: 61 SIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKI 119
S PPLNL SLE L LS CS LE FPEILGEM N+ L L PIKEL F FQNL L +
Sbjct: 2056 SFPPLNLTSLETLQLSGCSSLEYFPEILGEMENIKQLVLRDLPIKELQFSFQNLIGLQVL 2235
Query: 120 YPCNCGFVSLPTSNYAMSKLVECTIQ 145
Y +C V LP M +L + I+
Sbjct: 2236 YLWSCLIVELPCRLAMMPELFQLHIE 2313
>TC225609 UP|Q84ZV3 (Q84ZV3) R 4 protein, complete
Length = 2688
Score = 137 bits (346), Expect = 2e-33
Identities = 79/140 (56%), Positives = 87/140 (61%), Gaps = 1/140 (0%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
+ VL FD LTQIPDVS L NL E SF+ C +L+ V DSIG L +LK L C KL
Sbjct: 1882 LTVLKFDRCKFLTQIPDVSDLPNLRELSFEDCESLVAVDDSIGFLKKLKKLSAYGCRKLT 2061
Query: 61 SIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKI 119
S PPLNL SLE L LS CS LE FPEILGEM N+ L L IKELPF FQNLT L +
Sbjct: 2062 SFPPLNLTSLETLQLSSCSSLEYFPEILGEMENIRELRLTGLYIKELPFSFQNLTGLRLL 2241
Query: 120 YPCNCGFVSLPTSNYAMSKL 139
CG V LP S M +L
Sbjct: 2242 ALSGCGIVQLPCSLAMMPEL 2301
>TC225612 homologue to UP|Q84ZU8 (Q84ZU8) R 10 protein, complete
Length = 2706
Score = 137 bits (346), Expect = 2e-33
Identities = 77/142 (54%), Positives = 93/142 (65%), Gaps = 1/142 (0%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
+ VL FD+ LTQIPDVS L NL E SF+ C +L+ V DSIG L +LK L C KL+
Sbjct: 1888 LTVLKFDNCKFLTQIPDVSDLPNLRELSFEECESLVAVDDSIGFLNKLKKLSAYGCSKLK 2067
Query: 61 SIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKI 119
S PPLNL SL+ L+LS+CS LE FPEI+GEM N+ +L L PIKEL F FQNL L +
Sbjct: 2068 SFPPLNLTSLQTLELSQCSSLEYFPEIIGEMENIKHLFLYGLPIKELSFSFQNLIGLRWL 2247
Query: 120 YPCNCGFVSLPTSNYAMSKLVE 141
+CG V LP S M +L E
Sbjct: 2248 TLRSCGIVKLPCSLAMMPELFE 2313
>TC229101 disease resistance-like protein
Length = 2755
Score = 129 bits (323), Expect = 8e-31
Identities = 67/140 (47%), Positives = 90/140 (63%), Gaps = 1/140 (0%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
MRVLNFD LT+ PD+SG L+E S C NL+ +HDS+G L +L+ + C KL
Sbjct: 1213 MRVLNFDRCKFLTRTPDLSGAPILKELSSVFCENLVEIHDSVGFLDKLEIMNFESCSKLE 1392
Query: 61 SIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKI 119
+ PP+ L SLE ++LS CS L SFPEILG+M N+T+L+LEYT I +LP + L L +
Sbjct: 1393 TFPPIKLTSLESINLSHCSSLVSFPEILGKMENITHLSLEYTAISKLPNSIRELVRLQSL 1572
Query: 120 YPCNCGFVSLPTSNYAMSKL 139
NCG V LP+S + +L
Sbjct: 1573 ELHNCGMVQLPSSIVTLREL 1632
>TC215950 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidate
resistance protein KR1, partial (23%)
Length = 1891
Score = 125 bits (314), Expect = 9e-30
Identities = 65/112 (58%), Positives = 77/112 (68%)
Frame = +1
Query: 4 LNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP 63
L D LT+IPDVS L NLE SF C+NL +H S+GLLG+LK L C +L+S P
Sbjct: 517 LILDECDSLTEIPDVSCLSNLENLSFSECLNLFRIHHSVGLLGKLKILNAEGCPELKSFP 696
Query: 64 PLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT 115
PL L SLE LDLS CS LESFPEILG+M N+T L+L PI +LP F+NLT
Sbjct: 697 PLKLTSLESLDLSYCSSLESFPEILGKMENITELDLSECPITKLPPSFRNLT 852
>NP520295 disease resistance-like protein GS5-3 [Glycine max]
Length = 467
Score = 125 bits (313), Expect = 1e-29
Identities = 68/112 (60%), Positives = 75/112 (66%)
Frame = +1
Query: 3 VLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSI 62
+LNFD LT+IPDVS L NLEE SF C +L+ V DSIG L +LK L C KL S
Sbjct: 91 ILNFDQCEFLTKIPDVSDLPNLEELSFNWCESLVAVDDSIGFLNKLKKLSAYGCRKLTSF 270
Query: 63 PPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNL 114
PPLNL SLE L LS CS LE FPEILGEM N+ +L L IKEL F FQNL
Sbjct: 271 PPLNLTSLETLALSHCSSLEYFPEILGEMENIKHLFLYGLHIKELQFSFQNL 426
>TC207788 homologue to UP|Q8W2C0 (Q8W2C0) Functional candidate resistance
protein KR1, complete
Length = 3949
Score = 114 bits (285), Expect = 2e-26
Identities = 56/112 (50%), Positives = 76/112 (67%)
Frame = +1
Query: 4 LNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP 63
LNFD LT IPDVS + +L++ SF+ C NL +H S+G L +L+ L C +L++ P
Sbjct: 2014 LNFDSCQHLTLIPDVSCVPHLQKLSFKDCDNLYAIHPSVGFLEKLRILDAEGCSRLKNFP 2193
Query: 64 PLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT 115
P+ L SLE+L L C LE+FPEILG+M N+T L+LE TP+K+ FQNLT
Sbjct: 2194 PIKLTSLEQLKLGFCHSLENFPEILGKMENITELDLEQTPVKKFTLSFQNLT 2349
>TC215949 similar to UP|Q8W2C0 (Q8W2C0) Functional candidate resistance
protein KR1, partial (30%)
Length = 1805
Score = 113 bits (282), Expect = 5e-26
Identities = 61/112 (54%), Positives = 73/112 (64%)
Frame = +2
Query: 4 LNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP 63
L D LT+IPDVS L LE+ SF C NL T+H S+GLL +LK L C +L+S P
Sbjct: 755 LTLDECDSLTEIPDVSCLSKLEKLSFGNCRNLFTIHHSVGLLEKLKFLNAESCPELKSFP 934
Query: 64 PLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT 115
PL L SLE L LS CS LESFPEILG+M N+T L+ I +LP F+NLT
Sbjct: 935 PLKLTSLESLKLSYCSSLESFPEILGKMENMTQLSWTGCAITKLPPSFRNLT 1090
>TC221834 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidate resistance
protein KR1, partial (45%)
Length = 3472
Score = 111 bits (278), Expect = 1e-25
Identities = 67/141 (47%), Positives = 87/141 (61%), Gaps = 2/141 (1%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
M VL+FD LTQIPD+SG NL+E SF C NL+ +H+S+G L +LK++ C KL
Sbjct: 2155 MSVLSFDKFKFLTQIPDLSGTPNLKELSFVLCENLVEIHESVGFLDKLKSMNFEGCSKLE 2334
Query: 61 SIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELP--FPFQNLTLPK 118
+ PP+ L SLE ++LS CS L SFPEILG+M EYT I LP F ++L L +
Sbjct: 2335 TFPPIKLTSLESINLSYCSSLVSFPEILGKM--------EYTAISILP*LF*LRSLQLHR 2490
Query: 119 IYPCNCGFVSLPTSNYAMSKL 139
CG V LP S+ MS+L
Sbjct: 2491 -----CGMVQLP-SSIVMSEL 2535
>BM188061 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (10%)
Length = 427
Score = 110 bits (276), Expect = 2e-25
Identities = 59/108 (54%), Positives = 70/108 (64%)
Frame = +3
Query: 4 LNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP 63
L D LT+IPDVS L NLE SF+ C NL T+H S+GLL +LK L CC +L+S P
Sbjct: 102 LILDECDSLTEIPDVSCLSNLENLSFRKCRNLFTIHHSVGLLEKLKILDAECCPELKSFP 281
Query: 64 PLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPF 111
PL L SLE +L C LESFPEILG+M N+T L L PI +LP F
Sbjct: 282 PLKLTSLERFELWYCVSLESFPEILGKMENITQLCLYECPITKLPPSF 425
>BU091472 similar to GP|9965103|gb|A resistance protein LM6 {Glycine max},
partial (10%)
Length = 426
Score = 107 bits (268), Expect = 2e-24
Identities = 55/87 (63%), Positives = 64/87 (73%)
Frame = +1
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++VLNF+ LT+IPDVS L+NLEE SF C NLITVHDSIG L +LK L C KL
Sbjct: 166 LKVLNFEQCEFLTEIPDVSXLLNLEELSFNRCGNLITVHDSIGFLNKLKILGATRCRKLT 345
Query: 61 SIPPLNLASLEELDLSECSCLESFPEI 87
+ PPLNL SLE L+LS CS LE+FPEI
Sbjct: 346 TFPPLNLTSLERLELSCCSSLENFPEI 426
>TC233535 weakly similar to UP|Q6XZH4 (Q6XZH4) Nematode resistance-like
protein (Fragment), partial (4%)
Length = 741
Score = 101 bits (251), Expect = 2e-22
Identities = 62/147 (42%), Positives = 86/147 (58%), Gaps = 3/147 (2%)
Frame = +3
Query: 4 LNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP 63
++F L+++PD+SG+ +L C+NLI +HDS+G LG L+ L + C L+ IP
Sbjct: 246 MDFTDCEFLSEVPDISGIPDLRILYLDNCINLIKIHDSVGFLGNLEELTTIGCTSLKIIP 425
Query: 64 -PLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKIYP 121
LASL EL SEC L FPEIL E+ N+ LNL T I+ELPF NL L +
Sbjct: 426 SAFKLASLRELSFSECLRLVRFPEILCEIENLKYLNLWQTAIEELPFSIGNLRGLESLNL 605
Query: 122 CNCGFV-SLPTSNYAMSKLVECTIQAE 147
C + LP+S +A+ +L E IQA+
Sbjct: 606 MECARLDKLPSSIFALPRLQE--IQAD 680
>TC215948 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidate
resistance protein KR1, partial (19%)
Length = 1464
Score = 94.7 bits (234), Expect = 2e-20
Identities = 52/96 (54%), Positives = 61/96 (63%)
Frame = +2
Query: 20 GLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPLNLASLEELDLSECS 79
GL NLE SF C NL T+H S+GLL +LK L C KL+S L L SLE L+L C
Sbjct: 2 GLSNLENLSFGECRNLFTIHHSVGLLEKLKILDAQDCPKLKSFSSLKLTSLERLELWYCW 181
Query: 80 CLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT 115
LESF EILG+M N+T L L PI +LP F+NLT
Sbjct: 182 SLESFSEILGKMENITELFLTDCPITKLPPSFRNLT 289
>AW707276 similar to GP|9858483|gb|A resistance protein MG55 {Glycine max},
partial (36%)
Length = 571
Score = 72.4 bits (176), Expect = 9e-14
Identities = 39/111 (35%), Positives = 65/111 (58%), Gaps = 2/111 (1%)
Frame = +2
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
+++LN H LT P+ SGL LE+ + C ++ VH SIG L +L + + C L
Sbjct: 191 LKILNLSHSKYLTATPNFSGLPGLEKLILKDCPSMSKVHKSIGDLHKLVLINMKDCTSLT 370
Query: 61 SIP--PLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPF 109
++P +L S++ L+LS CS ++ E + +M ++T L EYT +K++PF
Sbjct: 371 NLPREMYHLKSVKTLNLSGCSEIDKL*EDILQMESLTTLIAEYTAVKQMPF 523
>BM094819 weakly similar to PIR|T47438|T47 disease resistance protein homolog
- Arabidopsis thaliana, partial (4%)
Length = 407
Score = 68.9 bits (167), Expect = 1e-12
Identities = 43/112 (38%), Positives = 60/112 (53%), Gaps = 4/112 (3%)
Frame = +3
Query: 32 CVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPP-LNLASLEELDLSECSCLESFPEILGE 90
C NL+ + SIG L +L TL C KL+ + + L SLE LDL +C CLE FPE+L +
Sbjct: 48 CSNLVKIDCSIGFLDKLLTLSAKGCSKLKILAHCIMLTSLEILDLGDCLCLEGFPEVLVK 227
Query: 91 MRNVTNLNLEYTPIKELPFPFQNLT---LPKIYPCNCGFVSLPTSNYAMSKL 139
M + + L+ T I LPF NL L + C + LP S + + K+
Sbjct: 228 MEKIREICLDNTAIGTLPFSIGNLVGLELLSLEQCK-RLIQLPGSIFTLPKV 380
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.142 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,547,150
Number of Sequences: 63676
Number of extensions: 121521
Number of successful extensions: 768
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 736
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 759
length of query: 164
length of database: 12,639,632
effective HSP length: 90
effective length of query: 74
effective length of database: 6,908,792
effective search space: 511250608
effective search space used: 511250608
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC145512.8


