

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.2 + phase: 0 /pseudo
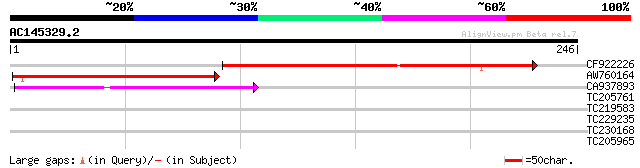
(246 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922226 108 2e-24
AW760164 similar to GP|11994422|dbj oxidoreductase short-chain ... 91 6e-19
CA937893 similar to GP|20805072|dbj retrovirus-related pol polyp... 54 7e-08
TC205761 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%) 31 0.45
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 29 1.7
TC229235 similar to UP|Q9LHR6 (Q9LHR6) Arabidopsis thaliana geno... 28 5.0
TC230168 28 5.0
TC205965 27 6.5
>CF922226
Length = 667
Score = 108 bits (270), Expect = 2e-24
Identities = 58/148 (39%), Positives = 91/148 (61%), Gaps = 11/148 (7%)
Frame = -3
Query: 93 MTKSLAHRQFLK*QLYLFRMVESKAIIEQLMEFNKILDDLENIEVQLEDEDKAILLFCAI 152
MTKSL +R + K LY F+M E +++ EQL FNK++ DLENI+V ++DED+A+LL C +
Sbjct: 665 MTKSLVNRLYXKQSLYSFKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYL 486
Query: 153 PKSFESFKDTMLYGKEGTVILEEVQAALRTKKLTKSKDLRVHEHGEGLSVL--------- 203
PKS+ FK+T+L+G++ +V L+EVQ AL +K+L + K+ + GEGL+
Sbjct: 485 PKSYSHFKETLLFGRD-SVSLDEVQTALNSKELNERKEKKSSASGEGLTARGKTFKKDSE 309
Query: 204 --RGNDGGRGNRRKSGNKSRSECFNCHK 229
+ + GN + C++C K
Sbjct: 308 FDKKKQKPENQKNGEGNIFKIRCYHCKK 225
>AW760164 similar to GP|11994422|dbj oxidoreductase short-chain
dehydrogenase/reductase family-like protein {Arabidopsis
thaliana}, partial (9%)
Length = 428
Score = 90.5 bits (223), Expect = 6e-19
Identities = 48/91 (52%), Positives = 65/91 (70%), Gaps = 1/91 (1%)
Frame = +3
Query: 2 MGS-KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVR 60
MGS K+++EKFTG NDFGL +KM A+L+QQ +AL GE L M+ +K ++ K
Sbjct: 75 MGSAKYEVEKFTGQNDFGLC*LKMRALLVQQGLVEALDGEIKLEKMMADGDKKALLQKAY 254
Query: 61 SAIVLCLGDKVLREVAKEPTAASIWAKLESL 91
+AI+L LGDKVLR+V+KE TA +W+KLE L
Sbjct: 255 NAIILSLGDKVLRQVSKETTAVGVWSKLEVL 347
>CA937893 similar to GP|20805072|dbj retrovirus-related pol polyprotein from
transposon TNT 1-94-like, partial (7%)
Length = 412
Score = 53.9 bits (128), Expect = 7e-08
Identities = 31/106 (29%), Positives = 57/106 (53%)
Frame = -2
Query: 3 GSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRSA 62
G+K+++ KF G +F LW+ +++ +L Q K L+ ++ ++ ++ +
Sbjct: 315 GAKFEVGKFDGTGNFRLWQKRVKDLLA*QGLLKVLRDSK--SNNTEALDWEEL*ERTATT 142
Query: 63 IVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLY 108
I LCL D+ L + + +W KLES YM KSL ++ +L +LY
Sbjct: 141 IRLCLVDEFLYHMMELAFPGEVWKKLESQYMLKSLTNKLYLMQKLY 4
>TC205761 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%)
Length = 1023
Score = 31.2 bits (69), Expect = 0.45
Identities = 13/26 (50%), Positives = 16/26 (61%)
Frame = -1
Query: 202 VLRGNDGGRGNRRKSGNKSRSECFNC 227
V R N GGRGN + GN++R C C
Sbjct: 228 VQRSNRGGRGNGDRHGNETRRTCLFC 151
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 29.3 bits (64), Expect = 1.7
Identities = 14/32 (43%), Positives = 15/32 (46%)
Frame = +2
Query: 196 HGEGLSVLRGNDGGRGNRRKSGNKSRSECFNC 227
HG+G G D GR RR G CFNC
Sbjct: 470 HGQGGG---GGDDGRNRRRGGGGGGGGGCFNC 556
>TC229235 similar to UP|Q9LHR6 (Q9LHR6) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K3D20, partial (78%)
Length = 1379
Score = 27.7 bits (60), Expect = 5.0
Identities = 15/45 (33%), Positives = 27/45 (59%)
Frame = +1
Query: 39 GEGLLPVTMSQVEKTDMVDKVRSAIVLCLGDKVLREVAKEPTAAS 83
GEG+L + + ++ D+ K+++ +V GDK +A+EP A S
Sbjct: 178 GEGILFIGNLRGKREDVFAKLQNQLVEVTGDKYNLFMAEEPNADS 312
>TC230168
Length = 863
Score = 27.7 bits (60), Expect = 5.0
Identities = 12/42 (28%), Positives = 23/42 (54%)
Frame = -3
Query: 21 KVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRSA 62
++ AV+IQ C+++LKG L T ++ T+ + +A
Sbjct: 441 RMSQNAVMIQALCQRSLKGSVALKYTSYPIQATEEITVTANA 316
>TC205965
Length = 1409
Score = 27.3 bits (59), Expect = 6.5
Identities = 11/22 (50%), Positives = 12/22 (54%)
Frame = +2
Query: 207 DGGRGNRRKSGNKSRSECFNCH 228
DG RGN GNKS + CH
Sbjct: 389 DGTRGNNSSDGNKSDNNSEKCH 454
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,966,743
Number of Sequences: 63676
Number of extensions: 90907
Number of successful extensions: 513
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 512
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 512
length of query: 246
length of database: 12,639,632
effective HSP length: 95
effective length of query: 151
effective length of database: 6,590,412
effective search space: 995152212
effective search space used: 995152212
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC145329.2


