

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.12 - phase: 0
(319 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
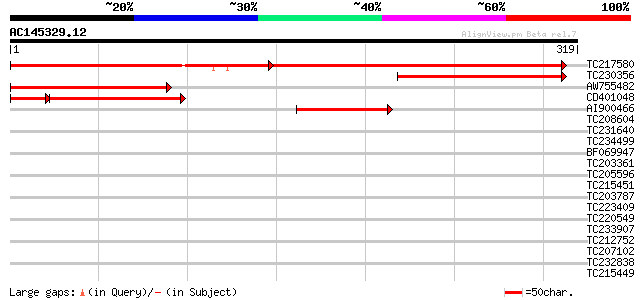
Score E
Sequences producing significant alignments: (bits) Value
TC217580 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (74%) 287 e-143
TC230356 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (34%) 168 3e-42
AW755482 similar to GP|28416701|gb| At4g02100 {Arabidopsis thali... 158 4e-39
CD401048 similar to GP|28416701|gb| At4g02100 {Arabidopsis thali... 132 1e-38
AI900466 94 6e-20
TC208604 38 0.007
TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated anti... 34 0.077
TC234499 weakly similar to UP|Q94A53 (Q94A53) At1g12830/F13K23_6... 33 0.17
BF069947 weakly similar to GP|24459855|emb disease resistance-li... 33 0.22
TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 33 0.22
TC205596 similar to GB|AAO63377.1|28950907|BT005313 At4g34530 {A... 32 0.29
TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembl... 32 0.38
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 32 0.38
TC223409 similar to UP|Q6T4P5 (Q6T4P5) Plasticity related gene 2... 32 0.38
TC220549 similar to UP|EOL1_ARATH (Q9ZQX6) ETO1-like protein 1 (... 32 0.50
TC233907 similar to UP|Q86NQ6 (Q86NQ6) RE01075p, partial (10%) 32 0.50
TC212752 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific... 32 0.50
TC207102 similar to UP|Q99MK4 (Q99MK4) AF9 (Fragment), partial (5%) 32 0.50
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 31 0.65
TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%) 31 0.85
>TC217580 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (74%)
Length = 2139
Score = 287 bits (735), Expect(2) = e-143
Identities = 140/167 (83%), Positives = 151/167 (89%)
Frame = +2
Query: 147 GLCKSCEKEGQWRYLVLGEACCHLGLMEDAMVLLQTGKRIASAAFRRESVCWSDDSFPLL 206
G CK+C+KEGQWRYLVLGEACCHLGLMEDAMVLLQTGKRIASAAFRRESVCWS+DSF +
Sbjct: 638 G*CKTCDKEGQWRYLVLGEACCHLGLMEDAMVLLQTGKRIASAAFRRESVCWSEDSFYVT 817
Query: 207 TIPLAGDTPNQQPTTPPRAPLNETESVTHLLSHIKFLLRRRAAALAALDAGLYSEAIRHF 266
IP +GD+ N P+TPPR L ++ESV LL HIKFLLRRRAAALAALDAGLYSEAIRHF
Sbjct: 818 NIPFSGDSTNAPPSTPPRTLLADSESVAQLLGHIKFLLRRRAAALAALDAGLYSEAIRHF 997
Query: 267 SKIVDGRRAAPQGFLAECYMHRASAHRSDGRIAESIADCNRTLSLDP 313
SKIVDGRR+APQ FLAECYMHRASAHRS GRIAESIADCNRTL+LDP
Sbjct: 998 SKIVDGRRSAPQSFLAECYMHRASAHRSAGRIAESIADCNRTLALDP 1138
Score = 240 bits (613), Expect(2) = e-143
Identities = 132/153 (86%), Positives = 141/153 (91%), Gaps = 5/153 (3%)
Frame = +1
Query: 1 MAVTTHSLTATEKKHWWLTNRKIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRL 60
MAVTT +L+ATEKKHWWLTNRKIVEKYIKDARSLIATQ+QSEI SALNL+DAALAISPR
Sbjct: 187 MAVTTRTLSATEKKHWWLTNRKIVEKYIKDARSLIATQDQSEIASALNLVDAALAISPRF 366
Query: 61 DQALELRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSS---SREG 117
DQALELRAR+LLYLRRFK+VADMLQDYIPSLRM N+D SS SSSSSSSSD+SS SREG
Sbjct: 367 DQALELRARALLYLRRFKEVADMLQDYIPSLRMGNDD-SSSSSSSSSSSDTSSQQLSREG 543
Query: 118 VKLL--SSDSPVRDQSFKCFSVSDLKKKVMAGL 148
VKLL SS+SPVRD SFKCFSVSDLKKKVMAGL
Sbjct: 544 VKLLSSSSESPVRDHSFKCFSVSDLKKKVMAGL 642
>TC230356 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (34%)
Length = 654
Score = 168 bits (426), Expect = 3e-42
Identities = 83/95 (87%), Positives = 89/95 (93%)
Frame = +3
Query: 219 PTTPPRAPLNETESVTHLLSHIKFLLRRRAAALAALDAGLYSEAIRHFSKIVDGRRAAPQ 278
PTTPPRAPL E+E+VT LL+HIK LLRRRAAALAALDAGL+SEAIRHFSKIVDGRR APQ
Sbjct: 6 PTTPPRAPLLESEAVTQLLAHIKLLLRRRAAALAALDAGLHSEAIRHFSKIVDGRRGAPQ 185
Query: 279 GFLAECYMHRASAHRSDGRIAESIADCNRTLSLDP 313
GFLAECYMHRASAH S GRIA+SIADCNRTL+LDP
Sbjct: 186 GFLAECYMHRASAHHSAGRIADSIADCNRTLALDP 290
>AW755482 similar to GP|28416701|gb| At4g02100 {Arabidopsis thaliana},
partial (14%)
Length = 433
Score = 158 bits (399), Expect = 4e-39
Identities = 81/91 (89%), Positives = 85/91 (93%)
Frame = +2
Query: 1 MAVTTHSLTATEKKHWWLTNRKIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRL 60
MAVTT + ATEKKHWWLTNRKIVEKY++DARSLIATQEQSEI SALNLLDAALAI PRL
Sbjct: 161 MAVTTPCINATEKKHWWLTNRKIVEKYMRDARSLIATQEQSEIGSALNLLDAALAIYPRL 340
Query: 61 DQALELRARSLLYLRRFKDVADMLQDYIPSL 91
D+ALELRAR LLYLRRFKDVADMLQDYIPSL
Sbjct: 341 DEALELRARCLLYLRRFKDVADMLQDYIPSL 433
>CD401048 similar to GP|28416701|gb| At4g02100 {Arabidopsis thaliana},
partial (14%)
Length = 642
Score = 132 bits (332), Expect(2) = 1e-38
Identities = 69/77 (89%), Positives = 73/77 (94%)
Frame = +2
Query: 23 IVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALELRARSLLYLRRFKDVAD 82
IVEKY++DARSLIATQEQSEI SALNLLDAALAI PRLD+ALELRARSLLYLRRFKDVAD
Sbjct: 410 IVEKYMRDARSLIATQEQSEIASALNLLDAALAIYPRLDEALELRARSLLYLRRFKDVAD 589
Query: 83 MLQDYIPSLRMTNEDPS 99
MLQDYIPSLRM+N D S
Sbjct: 590 MLQDYIPSLRMSNNDDS 640
Score = 45.4 bits (106), Expect(2) = 1e-38
Identities = 19/23 (82%), Positives = 20/23 (86%)
Frame = +1
Query: 1 MAVTTHSLTATEKKHWWLTNRKI 23
MAVTT L ATEKKHWWLTNRK+
Sbjct: 268 MAVTTPCLNATEKKHWWLTNRKV 336
>AI900466
Length = 163
Score = 94.4 bits (233), Expect = 6e-20
Identities = 44/54 (81%), Positives = 48/54 (88%)
Frame = +2
Query: 162 VLGEACCHLGLMEDAMVLLQTGKRIASAAFRRESVCWSDDSFPLLTIPLAGDTP 215
VLG+ACCHLGLMEDAMVLLQTGKR+A+AAFRRESVCWSDDSF L +GDTP
Sbjct: 2 VLGKACCHLGLMEDAMVLLQTGKRLATAAFRRESVCWSDDSFSLSNSLFSGDTP 163
>TC208604
Length = 1426
Score = 37.7 bits (86), Expect = 0.007
Identities = 15/33 (45%), Positives = 27/33 (81%)
Frame = +1
Query: 280 FLAECYMHRASAHRSDGRIAESIADCNRTLSLD 312
F+A C+ +RA+AH++ +IA++IADC+ ++LD
Sbjct: 40 FMAICFCNRAAAHQALDQIADAIADCSVAIALD 138
>TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated antigen, partial
(7%)
Length = 466
Score = 34.3 bits (77), Expect = 0.077
Identities = 22/51 (43%), Positives = 27/51 (52%)
Frame = -3
Query: 81 ADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQS 131
+ M+ PS ++ SS SSSSSSSS SSSS SS S V D +
Sbjct: 158 SSMITRTFPSSSSSSSFSSSSSSSSSSSSSSSSSSSSSSFSSSSSWVSDST 6
Score = 30.0 bits (66), Expect = 1.5
Identities = 19/38 (50%), Positives = 21/38 (55%)
Frame = -3
Query: 88 IPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDS 125
+ S +T PSS SSSS SSS SSSS SS S
Sbjct: 164 LSSSMITRTFPSSSSSSSFSSSSSSSSSSSSSSSSSSS 51
>TC234499 weakly similar to UP|Q94A53 (Q94A53) At1g12830/F13K23_6, partial
(26%)
Length = 894
Score = 33.1 bits (74), Expect = 0.17
Identities = 40/121 (33%), Positives = 51/121 (42%), Gaps = 4/121 (3%)
Frame = -2
Query: 22 KIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRL-DQALELRARSLLYLRRFKDV 80
K + ++ SL + S ++ A AA SP + L +R R L FK
Sbjct: 641 KTLTNLCRNQASLSSLSLSSSLMPAAEAAAAAFPESPSMRTPELAVRPRVLDGRMLFKST 462
Query: 81 ADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSS---DSPVRDQSFKCFSV 137
+ M SL ++ P S SSSSSSS SSSS LSS P R S FS
Sbjct: 461 S-MSGSSEKSLPSSSLSPPSSESSSSSSSSSSSSIIFPLPLSSRIIPFPFRSSSLSSFSP 285
Query: 138 S 138
S
Sbjct: 284 S 282
>BF069947 weakly similar to GP|24459855|emb disease resistance-like protein
{Coffea arabica}, partial (30%)
Length = 426
Score = 32.7 bits (73), Expect = 0.22
Identities = 23/66 (34%), Positives = 31/66 (46%), Gaps = 2/66 (3%)
Frame = -3
Query: 89 PSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLL--SSDSPVRDQSFKCFSVSDLKKKVMA 146
PS N SS SS SSSSS SSSS+ + ++ S R FK + +L +
Sbjct: 277 PSFSKNNSQASSSSSFSSSSSSSSSSKGNIVIILYGWQSICRHTCFKRVVIINLMPFTLR 98
Query: 147 GLCKSC 152
G+ C
Sbjct: 97 GIPTFC 80
>TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 1090
Score = 32.7 bits (73), Expect = 0.22
Identities = 19/33 (57%), Positives = 21/33 (63%)
Frame = +3
Query: 94 TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSP 126
++ PSS SSSSSSSS SSSS SS SP
Sbjct: 324 SSSPPSSSSSSSSSSSSSSSS*SSPSSSSSSSP 422
Score = 28.9 bits (63), Expect = 3.2
Identities = 19/39 (48%), Positives = 19/39 (48%)
Frame = +3
Query: 98 PSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFS 136
PSS SSS SSS SSSS S SP S FS
Sbjct: 312 PSSSSSSPPSSSSSSSSSSSSSSSS*SSPSSSSSSSPFS 428
Score = 27.7 bits (60), Expect = 7.2
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +3
Query: 94 TNEDPSSGSSSSSSSSDSSSS 114
++ P S SSSSSSSS SSSS
Sbjct: 321 SSSSPPSSSSSSSSSSSSSSS 383
>TC205596 similar to GB|AAO63377.1|28950907|BT005313 At4g34530 {Arabidopsis
thaliana;} , partial (28%)
Length = 1029
Score = 32.3 bits (72), Expect = 0.29
Identities = 21/68 (30%), Positives = 31/68 (44%)
Frame = +3
Query: 58 PRLDQALELRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREG 117
P + L + L R ++ Q++ P SSSSSSSS SSS +G
Sbjct: 99 PTMLHCLNTSCSDITVLERQREATIKCQNHQPPYLTDFNAVFPSSSSSSSSSSSSSHSQG 278
Query: 118 VKLLSSDS 125
+ ++ SDS
Sbjct: 279 LLMMCSDS 302
>TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembly protein,
partial (20%)
Length = 777
Score = 32.0 bits (71), Expect = 0.38
Identities = 22/50 (44%), Positives = 28/50 (56%)
Frame = -1
Query: 77 FKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSP 126
FKD+ + ++ L ++ SS SSSSSSSS SSSS SS SP
Sbjct: 225 FKDI-NCYTFFLVLLSSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSRSP 79
Score = 30.0 bits (66), Expect = 1.5
Identities = 18/33 (54%), Positives = 20/33 (60%)
Frame = -1
Query: 99 SSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQS 131
SS SSSSSSSS SSSS +SS S +S
Sbjct: 180 SSSSSSSSSSSSSSSSSSSSSSISSSSSSSSRS 82
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 32.0 bits (71), Expect = 0.38
Identities = 18/33 (54%), Positives = 21/33 (63%)
Frame = +3
Query: 94 TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSP 126
++ P+S SSSSSSSS SSSS SS SP
Sbjct: 357 SSSSPASSSSSSSSSSSSSSS*SSPSSSSSSSP 455
>TC223409 similar to UP|Q6T4P5 (Q6T4P5) Plasticity related gene 2a, partial
(3%)
Length = 439
Score = 32.0 bits (71), Expect = 0.38
Identities = 18/24 (75%), Positives = 19/24 (79%)
Frame = -3
Query: 99 SSGSSSSSSSSDSSSSREGVKLLS 122
SS SSSSSSSS SSSS +KLLS
Sbjct: 89 SSSSSSSSSSSSSSSSPTNLKLLS 18
Score = 27.7 bits (60), Expect = 7.2
Identities = 17/36 (47%), Positives = 21/36 (58%)
Frame = -3
Query: 91 LRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSP 126
L + + + G+SSSSSSS SSSS SS SP
Sbjct: 131 LNLISSEEDKGASSSSSSSSSSSSSS-----SSSSP 39
>TC220549 similar to UP|EOL1_ARATH (Q9ZQX6) ETO1-like protein 1 (Ethylene
overproducer 1-like protein 1), partial (26%)
Length = 876
Score = 31.6 bits (70), Expect = 0.50
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 2/87 (2%)
Frame = +3
Query: 229 ETESVTHLLSHIKFLLRRRAAALAALDAGLYSEAIRHFSKIVDGRRAAPQGFLAECYMH- 287
+ E VT L + R RAA L +D+ EAI S+ + F A+ ++
Sbjct: 405 DLEMVTRLDPLRVYPYRYRAAVL--MDSHKEEEAIAELSRAI--------AFKADLHLLH 554
Query: 288 -RASAHRSDGRIAESIADCNRTLSLDP 313
RA+ H G + +I DC LS+DP
Sbjct: 555 LRAAFHEHKGDVLGAIRDCRAALSVDP 635
>TC233907 similar to UP|Q86NQ6 (Q86NQ6) RE01075p, partial (10%)
Length = 845
Score = 31.6 bits (70), Expect = 0.50
Identities = 36/104 (34%), Positives = 49/104 (46%)
Frame = -1
Query: 54 LAISPRLDQALELRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSS 113
LAIS R ++ LR RS L V + Y L SS SSSSSSSS SSS
Sbjct: 368 LAISERDKKSPMLRHRSSL-------VEPLTLKYSSLL-----SSSSSSSSSSSSSSSSS 225
Query: 114 SREGVKLLSSDSPVRDQSFKCFSVSDLKKKVMAGLCKSCEKEGQ 157
S V+ +SS ++SD K+K L ++ ++ G+
Sbjct: 224 SNSDVE*ISS----YKVGVAIITMSDDKRKGSCVLAEAEDESGE 105
>TC212752 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific protein T2
precursor, partial (35%)
Length = 533
Score = 31.6 bits (70), Expect = 0.50
Identities = 22/54 (40%), Positives = 25/54 (45%), Gaps = 7/54 (12%)
Frame = -2
Query: 92 RMTNED-------PSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVS 138
R TN D PS S +SSSSS SSSS + SS S CF +S
Sbjct: 178 RHTNSDSLSPSSSPSQSSHTSSSSSSSSSSSSSLSSSSSSILCGSSSSSCFCLS 17
>TC207102 similar to UP|Q99MK4 (Q99MK4) AF9 (Fragment), partial (5%)
Length = 1414
Score = 31.6 bits (70), Expect = 0.50
Identities = 18/52 (34%), Positives = 28/52 (53%)
Frame = +2
Query: 79 DVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQ 130
+ A+ L+D + +L + +S SSS SSSS+S S +SDS D+
Sbjct: 986 NAAEALEDQVNALGRDEQTSTSSSSSGSSSSESGSESGSGSSSNSDSEGSDE 1141
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 31.2 bits (69), Expect = 0.65
Identities = 24/62 (38%), Positives = 28/62 (44%)
Frame = -3
Query: 90 SLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDLKKKVMAGLC 149
S+ ++ P S SSSSSSSS SS S SS SP R S S S +G
Sbjct: 579 SVPKSSSSPCSSSSSSSSSSSSSESSSESLSSSSGSPPRWTSPSSSSSSSSSSSPPSGAS 400
Query: 150 KS 151
S
Sbjct: 399 SS 394
Score = 30.8 bits (68), Expect = 0.85
Identities = 21/61 (34%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Frame = -3
Query: 94 TNEDPSSGSSSSSS--SSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDLKKKVMAGLCKS 151
++ P SG+SSSSS SS SSSS SS + S +S S K + +
Sbjct: 429 SSSSPPSGASSSSSSLSSPSSSSSSSSSSSSSSASSTTSSTDSYSSSSSSTKTLTSFSST 250
Query: 152 C 152
C
Sbjct: 249 C 247
Score = 28.1 bits (61), Expect = 5.5
Identities = 19/41 (46%), Positives = 20/41 (48%)
Frame = -3
Query: 98 PSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVS 138
PSS SSSSSSSS S + LSS S S S S
Sbjct: 453 PSSSSSSSSSSSPPSGASSSSSSLSSPSSSSSSSSSSSSSS 331
Score = 27.7 bits (60), Expect = 7.2
Identities = 16/36 (44%), Positives = 21/36 (57%)
Frame = -3
Query: 90 SLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDS 125
S +++ SS SSSSSSSS +SS+ SS S
Sbjct: 393 SSSLSSPSSSSSSSSSSSSSSASSTTSSTDSYSSSS 286
>TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%)
Length = 1495
Score = 30.8 bits (68), Expect = 0.85
Identities = 19/40 (47%), Positives = 23/40 (57%)
Frame = -2
Query: 87 YIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSP 126
++ L ++ SS SSSSSSSS SSSS SS SP
Sbjct: 1152 FLVLLSSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSRSP 1033
Score = 30.0 bits (66), Expect = 1.5
Identities = 18/33 (54%), Positives = 20/33 (60%)
Frame = -2
Query: 99 SSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQS 131
SS SSSSSSSS SSSS +SS S +S
Sbjct: 1134 SSSSSSSSSSSSSSSSSSSSSSISSSSSSSSRS 1036
Score = 28.5 bits (62), Expect = 4.2
Identities = 16/29 (55%), Positives = 19/29 (65%)
Frame = -2
Query: 86 DYIPSLRMTNEDPSSGSSSSSSSSDSSSS 114
D+ L ++ SS SSSSSSSS SSSS
Sbjct: 1158 DFFLVLLSSSSSSSSSSSSSSSSSSSSSS 1072
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.130 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,945,919
Number of Sequences: 63676
Number of extensions: 232485
Number of successful extensions: 4986
Number of sequences better than 10.0: 138
Number of HSP's better than 10.0 without gapping: 2326
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3478
length of query: 319
length of database: 12,639,632
effective HSP length: 97
effective length of query: 222
effective length of database: 6,463,060
effective search space: 1434799320
effective search space used: 1434799320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC145329.12


