

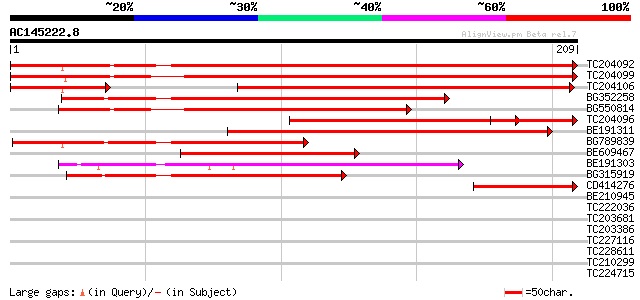
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.8 + phase: 0
(209 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204092 homologue to UP|PSAD_NICSY (P29302) Photosystem I react... 348 8e-97
TC204099 homologue to UP|PSAD_LYCES (P12372) Photosystem I react... 343 2e-95
TC204106 similar to UP|PSAD_NICSY (P29302) Photosystem I reactio... 209 7e-55
BG352258 similar to SP|P29302|PSAD_ Photosystem I reaction cente... 160 5e-40
BG550814 similar to SP|P32869|PSAD Photosystem I reaction center... 156 6e-39
TC204096 homologue to UP|PSAD_CUCSA (P32869) Photosystem I react... 154 2e-38
BE191311 120 6e-28
BG789839 similar to SP|P29302|PSAD_ Photosystem I reaction cente... 117 5e-27
BE609467 similar to SP|P29302|PSAD_ Photosystem I reaction cente... 106 7e-24
BE191303 91 3e-19
BG315919 weakly similar to SP|P36213|PSAD_ Photosystem I reactio... 88 2e-18
CD414276 homologue to GP|13877126|gb| ribosomal protein L32 {Mer... 77 4e-15
BE210945 36 0.014
TC222036 similar to PIR|E96668|E96668 protein F1N19.3 [imported]... 33 0.093
TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 33 0.093
TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 32 0.16
TC227116 similar to UP|WAS2_HUMAN (Q9Y6W5) Wiskott-Aldrich syndr... 32 0.27
TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxida... 31 0.35
TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor T... 31 0.35
TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%) 31 0.46
>TC204092 homologue to UP|PSAD_NICSY (P29302) Photosystem I reaction center
subunit II, chloroplast precursor (Photosystem I 20 kDa
subunit) (PSI-D), partial (85%)
Length = 934
Score = 348 bits (894), Expect = 8e-97
Identities = 178/216 (82%), Positives = 189/216 (87%), Gaps = 7/216 (3%)
Frame = +3
Query: 1 MAMATQASLFTPPLSSPK-------PWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAA 53
MAMATQASL TPPLS K PWKQ S+LSF S KP+KF+ +T + +AADE TEA
Sbjct: 129 MAMATQASLLTPPLSGLKASDRASVPWKQNSSLSFSSPKPLKFS-RTIRAAAADETTEAP 305
Query: 54 SVTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEM 113
+ + AAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITW+SPKEQIFEM
Sbjct: 306 A-----KVEAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWDSPKEQIFEM 470
Query: 114 PTGGAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYP 173
PTGGAAIMREGPNLLKLARKEQCLALG RLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYP
Sbjct: 471 PTGGAAIMREGPNLLKLARKEQCLALGTRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYP 650
Query: 174 EKVNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
EKVN GRQGVGQNFRSIGKNVSPIEVKFTGKQP+D+
Sbjct: 651 EKVNAGRQGVGQNFRSIGKNVSPIEVKFTGKQPYDL 758
>TC204099 homologue to UP|PSAD_LYCES (P12372) Photosystem I reaction center
subunit II, chloroplast precursor (Photosystem I 20 kDa
subunit) (PSI-D), partial (80%)
Length = 953
Score = 343 bits (881), Expect = 2e-95
Identities = 175/216 (81%), Positives = 185/216 (85%), Gaps = 7/216 (3%)
Frame = +2
Query: 1 MAMATQASLFTPPLSSPKP-------WKQPSTLSFISLKPIKFTTKTTKISAADEKTEAA 53
MAMATQASLFTPPLS KP WKQ STLSF + KP+KF+ +T + +A D+ TEA
Sbjct: 128 MAMATQASLFTPPLSGLKPSDRATAPWKQSSTLSFSTSKPLKFS-RTIRAAATDQTTEA- 301
Query: 54 SVTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEM 113
PVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITW+SPKEQIFEM
Sbjct: 302 -----------PVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWDSPKEQIFEM 448
Query: 114 PTGGAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYP 173
PTGGAAIMREGPNLLKLARKEQCLALG RLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYP
Sbjct: 449 PTGGAAIMREGPNLLKLARKEQCLALGTRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYP 628
Query: 174 EKVNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
EKVN GRQGVGQNFRSIGKNVSPIEVKFTGKQP+D+
Sbjct: 629 EKVNAGRQGVGQNFRSIGKNVSPIEVKFTGKQPYDL 736
>TC204106 similar to UP|PSAD_NICSY (P29302) Photosystem I reaction center
subunit II, chloroplast precursor (Photosystem I 20 kDa
subunit) (PSI-D), partial (74%)
Length = 536
Score = 209 bits (532), Expect = 7e-55
Identities = 97/124 (78%), Positives = 111/124 (89%)
Frame = +3
Query: 85 STGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGNRLR 144
STGGLLRKAQVEEFYVITW+SPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALG +L+
Sbjct: 165 STGGLLRKAQVEEFYVITWDSPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGTKLK 344
Query: 145 SKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGKNVSPIEVKFTGK 204
SKYKIKYQ Y++FPN EVQYLHP++ +YP K+NP RQ GQN RSI N++PI++K T K
Sbjct: 345 SKYKIKYQVYKIFPNREVQYLHPRNNIYP*KINPERQRFGQNLRSINNNINPIQLKGTSK 524
Query: 205 QPFD 208
QP+D
Sbjct: 525 QPYD 536
Score = 54.7 bits (130), Expect = 3e-08
Identities = 29/44 (65%), Positives = 32/44 (71%), Gaps = 7/44 (15%)
Frame = +1
Query: 1 MAMATQASLFTPPLSSPK-------PWKQPSTLSFISLKPIKFT 37
MAMATQASLFTPPLS K PWKQ STLSF + KP+KF+
Sbjct: 19 MAMATQASLFTPPLSGLKPSDRATAPWKQSSTLSFSTSKPLKFS 150
>BG352258 similar to SP|P29302|PSAD_ Photosystem I reaction center subunit II
chloroplast precursor (Photosystem I 20 kDa subunit),
partial (48%)
Length = 412
Score = 160 bits (404), Expect = 5e-40
Identities = 88/143 (61%), Positives = 98/143 (67%)
Frame = +2
Query: 20 WKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPS 79
WKQ TL L+ ++ K +A DE TE + + AAP G P +L+PN
Sbjct: 2 WKQKYTLYLFKLEALR-VVKNVGSAAVDETTEVPA-----KVEAAPRGIMPRQLEPNNSC 163
Query: 80 PIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLAL 139
I GSTGGLLRKAQVEE YV+T +S KEQIFEMPTGGA IMREG NLLKLARKE CLAL
Sbjct: 164 LILWGSTGGLLRKAQVEELYVLTGDSSKEQIFEMPTGGAVIMREGANLLKLARKEHCLAL 343
Query: 140 GNRLRSKYKIKYQFYRVFPNGEV 162
G RLRS YKIKY FYRVF NGEV
Sbjct: 344 GTRLRS*YKIKYYFYRVFHNGEV 412
>BG550814 similar to SP|P32869|PSAD Photosystem I reaction center subunit II
chloroplast precursor (Photosystem I 20 kDa subunit),
partial (48%)
Length = 365
Score = 156 bits (395), Expect = 6e-39
Identities = 82/130 (63%), Positives = 91/130 (69%)
Frame = +3
Query: 19 PWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTP 78
PWKQ STLSF + KP+KF+ +T + +A D+ TEA PVG TPP LDPNTP
Sbjct: 15 PWKQSSTLSFSTSKPLKFS-RTIRAAATDQTTEA------------PVGVTPPVLDPNTP 155
Query: 79 SPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLA 138
PIFGG GGLLR AQVEEFYVITW+SPKEQ + P GGA IMREGPNLLK RKEQC
Sbjct: 156 XPIFGGGGGGLLRTAQVEEFYVITWDSPKEQK*KCPLGGAXIMREGPNLLKXXRKEQCWX 335
Query: 139 LGNRLRSKYK 148
G RLR K K
Sbjct: 336 FGTRLRVKDK 365
>TC204096 homologue to UP|PSAD_CUCSA (P32869) Photosystem I reaction center
subunit II, chloroplast precursor (Photosystem I 20 kDa
subunit) (PSI-D) (PS I subunit 5), partial (50%)
Length = 1054
Score = 154 bits (390), Expect = 2e-38
Identities = 75/85 (88%), Positives = 77/85 (90%)
Frame = +1
Query: 104 ESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQ 163
+SPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALG RLRSKYKIKYQF RVFPNGEVQ
Sbjct: 619 DSPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGTRLRSKYKIKYQF*RVFPNGEVQ 798
Query: 164 YLHPKDGVYPEKVNPGRQGVGQNFR 188
YLHPKDGVYPEKVN GRQ G +
Sbjct: 799 YLHPKDGVYPEKVNAGRQRGGSKLQ 873
Score = 60.8 bits (146), Expect = 4e-10
Identities = 27/32 (84%), Positives = 30/32 (93%)
Frame = +2
Query: 178 PGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
P +GVGQNFRSIGKNVSPIEVKFTGKQP+D+
Sbjct: 842 PDAKGVGQNFRSIGKNVSPIEVKFTGKQPYDL 937
>BE191311
Length = 397
Score = 120 bits (300), Expect = 6e-28
Identities = 63/120 (52%), Positives = 80/120 (66%)
Frame = +2
Query: 81 IFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALG 140
IF T LL KA VEE YVI+ + KE IF +P+ A ++ EGPNL+KL K+Q L +
Sbjct: 38 IFLCITAELLHKAHVEETYVISLD*HKEHIFVLPSDSAVMITEGPNLVKLDMKDQSLTVA 217
Query: 141 NRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGKNVSPIEVK 200
L SKYKIK FYRV PNG+V Y +P DGV PE + R G GQN++SIG NV P+++K
Sbjct: 218 TMLMSKYKIKSHFYRVCPNGDVHYSYPYDGVNPETADARRLGGGQNYQSIGNNVIPMKLK 397
>BG789839 similar to SP|P29302|PSAD_ Photosystem I reaction center subunit II
chloroplast precursor (Photosystem I 20 kDa subunit),
partial (36%)
Length = 331
Score = 117 bits (292), Expect = 5e-27
Identities = 69/116 (59%), Positives = 76/116 (65%), Gaps = 7/116 (6%)
Frame = +2
Query: 2 AMATQASLFTPPLSSPK-------PWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAAS 54
AMATQA L TPPLS K PWKQ S+LSF S P++ +T AAD EA +
Sbjct: 2 AMATQAXLLTPPLSGLKASDRASVPWKQNSSLSFXSPNPLR-GGRTIFFFAAD*YPEAPA 178
Query: 55 VTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQI 110
+ AAP FTPPELDPNTPSPI GG T GLLRKAQVEE YVITW+ PKEQI
Sbjct: 179 -----KGEAAPAPFTPPELDPNTPSPIXGGRTRGLLRKAQVEEXYVITWDLPKEQI 331
>BE609467 similar to SP|P29302|PSAD_ Photosystem I reaction center subunit II
chloroplast precursor (Photosystem I 20 kDa subunit),
partial (26%)
Length = 199
Score = 106 bits (265), Expect = 7e-24
Identities = 49/66 (74%), Positives = 55/66 (83%)
Frame = +2
Query: 64 APVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMRE 123
APVGFTPPELDP PSPIFGGSTGGLLRKAQVEE V+TW SPKE IF++PTGGA ++
Sbjct: 2 APVGFTPPELDPPGPSPIFGGSTGGLLRKAQVEELDVLTWHSPKELIFDVPTGGAGVVTV 181
Query: 124 GPNLLK 129
GP L +
Sbjct: 182 GPRLFR 199
>BE191303
Length = 520
Score = 91.3 bits (225), Expect = 3e-19
Identities = 65/153 (42%), Positives = 87/153 (56%), Gaps = 4/153 (2%)
Frame = +2
Query: 19 PWKQPSTLSFISL--KPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPE-LDP 75
PW +T S+ SL P+K+ T+ S A + A + VG+ P + L
Sbjct: 74 PWNA*NT-SYPSLTESPVKY*HNRTEHSTAYKHHMRAP---SDSLMTDLVGYHPTQSLTS 241
Query: 76 NTPSPI-FGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKE 134
NT P F LL KA VEE YVI+ + K+ I E+P+GGAA++ EG NL+KLARK+
Sbjct: 242 NTNFPTSFWV*RRELLHKAHVEETYVISLD*HKKHILELPSGGAAMLTEGSNLVKLARKD 421
Query: 135 QCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHP 167
Q LA+ LRSK KIKY F +V+PNGE + P
Sbjct: 422 QSLAVSTMLRSKNKIKYHFLQVWPNGEGHWFAP 520
>BG315919 weakly similar to SP|P36213|PSAD_ Photosystem I reaction center
subunit II chloroplast precursor (Photosystem I 20 kDa
subunit), partial (30%)
Length = 292
Score = 88.2 bits (217), Expect = 2e-18
Identities = 50/103 (48%), Positives = 68/103 (65%)
Frame = +2
Query: 22 QPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSPI 81
Q TLS+ S +P+ + +K+ + +A D T+A + + AAP PP+LDPN P+
Sbjct: 2 QKYTLSYSSREPL-WASKSRRSAAVD*TTQAPA-----KVEAAPRWIIPPQLDPNNTCPM 163
Query: 82 FGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREG 124
G+T GLLRKAQ EE YV+T +S KE+I+EMPTG AIMREG
Sbjct: 164 LRGTTSGLLRKAQEEESYVMT*DSSKEKIYEMPTGMGAIMREG 292
>CD414276 homologue to GP|13877126|gb| ribosomal protein L32 {Mercurialis
annua}, partial (31%)
Length = 599
Score = 77.4 bits (189), Expect = 4e-15
Identities = 35/38 (92%), Positives = 37/38 (97%)
Frame = -1
Query: 172 YPEKVNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
YPEKVN GRQGVGQNFRSIGKNVSPIEVKFTGKQP+D+
Sbjct: 590 YPEKVNAGRQGVGQNFRSIGKNVSPIEVKFTGKQPYDL 477
>BE210945
Length = 416
Score = 35.8 bits (81), Expect = 0.014
Identities = 21/70 (30%), Positives = 32/70 (45%)
Frame = +2
Query: 12 PPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPP 71
PP+ +P P S + S P TT T K+ + S K P +PV + PP
Sbjct: 161 PPVKAPTP---ASPVKSPSYPPGSVTTPTVKVPPPPQ-----SPVVKPPTPTSPVVYPPP 316
Query: 72 ELDPNTPSPI 81
+ P+ P+P+
Sbjct: 317 PVAPSPPAPV 346
>TC222036 similar to PIR|E96668|E96668 protein F1N19.3 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(48%)
Length = 622
Score = 33.1 bits (74), Expect = 0.093
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 2/79 (2%)
Frame = +1
Query: 4 ATQASLFTPPLSSPKPWKQPST--LSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEA 61
A SL +PPL++P PST L ++ P TT + S +++ + S +
Sbjct: 163 APSRSLPSPPLTAPSTSPSPSTPPLGTLTEAPSPTTTARSSFSTTADRSASCSFRPERSP 342
Query: 62 PAAPVGFTPPELDPNTPSP 80
PA + PP P +P P
Sbjct: 343 PAGLSTWPPP--SPFSPFP 393
>TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 913
Score = 33.1 bits (74), Expect = 0.093
Identities = 28/93 (30%), Positives = 38/93 (40%), Gaps = 9/93 (9%)
Frame = +3
Query: 2 AMATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEA 61
A + A+ TPP SSP PS + + P+ T +A +VT
Sbjct: 207 ASSPNAATATPPASSPTVASPPSKAA--APAPVATPPAATPPAATPPAATPPAVTPVSSP 380
Query: 62 PA--------APVGFT-PPELDPNTPSPIFGGS 85
PA APV + PP L P TP+P+ S
Sbjct: 381 PAPVPVSSPPAPVPVSSPPALAPTTPAPVVAPS 479
>TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 649
Score = 32.3 bits (72), Expect = 0.16
Identities = 28/95 (29%), Positives = 36/95 (37%), Gaps = 11/95 (11%)
Frame = +1
Query: 2 AMATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKIS-AADEKTEAASVTTKEE 60
A + A+ TPP+SSP PS + P TT AA T
Sbjct: 181 ASSPNAATATPPVSSPTVASPPSKAA----APAPATTPPVATPPAATPPAATPPAVTPVS 348
Query: 61 APAAPVGFT----------PPELDPNTPSPIFGGS 85
+P APV + PP L P TP+P+ S
Sbjct: 349 SPPAPVPVSSPPAPVPVSSPPALAPTTPAPVVAPS 453
>TC227116 similar to UP|WAS2_HUMAN (Q9Y6W5) Wiskott-Aldrich syndrome protein
family member 2 (WASP-family protein member 2)
(Verprolin homology domain-containing protein 2),
partial (5%)
Length = 1154
Score = 31.6 bits (70), Expect = 0.27
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Frame = +3
Query: 12 PPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTT--KEEAPAAPVGFT 69
PPLSS + P++ S + +P TT S + KT + S T + AP+ P
Sbjct: 126 PPLSSART-ATPTSSSSWTPQPSPKTTPNPPSSTSFSKTLSYSSTPLPQNPAPSPPSTSP 302
Query: 70 PPELDPNTPSPIF 82
PP P+TP+ F
Sbjct: 303 PPSSPPSTPTASF 341
>TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26a
{Arabidopsis thaliana;} , partial (49%)
Length = 731
Score = 31.2 bits (69), Expect = 0.35
Identities = 21/77 (27%), Positives = 34/77 (43%)
Frame = +3
Query: 4 ATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPA 63
+T +S PPLSSP P P+ + P T + +A + +A+ T + +P
Sbjct: 321 STSSSAPRPPLSSPAPTPSPAP----TFSPPPPATSSPCSAAPSSPSSSAAATAEPPSPP 488
Query: 64 APVGFTPPELDPNTPSP 80
+PP P+ SP
Sbjct: 489 PYPPTSPPPPCPSLKSP 539
Score = 28.1 bits (61), Expect = 3.0
Identities = 21/74 (28%), Positives = 31/74 (41%), Gaps = 2/74 (2%)
Frame = +3
Query: 9 LFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKT--EAASVTTKEEAPAAPV 66
LF+ P + PW TL S + ++ ++ A A+S TT ++P
Sbjct: 63 LFSAPPTRDSPWTSTRTLVRSSARS*GTPSQASRSPAPPPPPPRSASSSTTA----SSPT 230
Query: 67 GFTPPELDPNTPSP 80
TPP P PSP
Sbjct: 231 AVTPPSSSPPRPSP 272
>TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 1615
Score = 31.2 bits (69), Expect = 0.35
Identities = 22/79 (27%), Positives = 34/79 (42%), Gaps = 1/79 (1%)
Frame = +2
Query: 2 AMATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEA 61
A + ++ PP P PW P + ++ P+K TT++T AAS +T +
Sbjct: 257 APSATSTTVRPPSPPPSPWPSPPS----AIVPLKNTTRST---PPPRSAPAASPSTPPPS 415
Query: 62 PAAP-VGFTPPELDPNTPS 79
P TP P TP+
Sbjct: 416 NTRPRTATTPTSTAPATPT 472
>TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%)
Length = 1270
Score = 30.8 bits (68), Expect = 0.46
Identities = 24/77 (31%), Positives = 38/77 (49%)
Frame = +2
Query: 13 PLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPE 72
P SSP P P++ + S P + + SAA + AAS T+ +P++ +PP
Sbjct: 704 PSSSPSP-SAPTSPAATSTPP----SPSAPSSAATSPSSAASSTS---SPSSSAPSSPPS 859
Query: 73 LDPNTPSPIFGGSTGGL 89
P++P P+F S L
Sbjct: 860 SWPSSPPPLFQHSDSPL 910
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,056,837
Number of Sequences: 63676
Number of extensions: 127531
Number of successful extensions: 1016
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 964
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 999
length of query: 209
length of database: 12,639,632
effective HSP length: 93
effective length of query: 116
effective length of database: 6,717,764
effective search space: 779260624
effective search space used: 779260624
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC145222.8


