

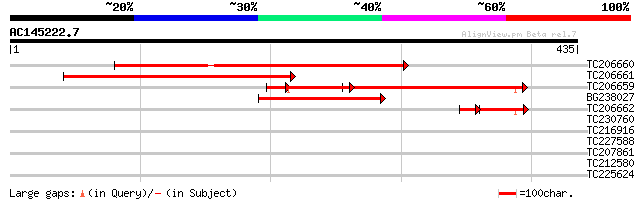
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.7 - phase: 0
(435 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206660 similar to UP|O23033 (O23033) YUP8H12.2 protein, partia... 422 e-118
TC206661 331 4e-91
TC206659 similar to UP|O23033 (O23033) YUP8H12.2 protein, partia... 226 5e-86
BG238027 158 4e-39
TC206662 similar to UP|O23033 (O23033) YUP8H12.2 protein, partia... 44 2e-09
TC230760 weakly similar to UP|Q9LVG7 (Q9LVG7) Ankyrin-like prote... 30 2.1
TC216916 similar to UP|Q9ARE3 (Q9ARE3) ZF-HD homeobox protein, p... 30 2.1
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 29 3.6
TC207861 similar to UP|MPK7_ARATH (Q39027) Mitogen-activated pro... 28 6.2
TC212580 28 8.1
TC225624 similar to UP|Q7Y240 (Q7Y240) Thioredoxin peroxidase 1,... 28 8.1
>TC206660 similar to UP|O23033 (O23033) YUP8H12.2 protein, partial (47%)
Length = 666
Score = 422 bits (1085), Expect = e-118
Identities = 204/226 (90%), Positives = 216/226 (95%)
Frame = +1
Query: 81 ALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLKSAPYDTIQLNRGPSWLDKN 140
ALGVASSIGLGSGLHTFVLYLGPHIALFTI A+QCGRVDLKSAPYDTIQL RGPSWLDK+
Sbjct: 1 ALGVASSIGLGSGLHTFVLYLGPHIALFTIGAVQCGRVDLKSAPYDTIQLKRGPSWLDKD 180
Query: 141 CSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGELPPYFISRAARLSGSRMDA 200
CS+FGPPLFQS+ VPLSSILPQVQ+EA+LWG+GTAIGELPPYFISRAARLSG R+DA
Sbjct: 181 CSEFGPPLFQSQ----VPLSSILPQVQLEAILWGIGTAIGELPPYFISRAARLSGGRVDA 348
Query: 201 MEELDSEDKGVLNQIKCWFFSHTQHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFWNFF 260
MEELDSEDK VL++IKCWF SH+QHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFW FF
Sbjct: 349 MEELDSEDKRVLSRIKCWFLSHSQHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFWKFF 528
Query: 261 LATLIGKAIIKTHIQTVFIISVCNNQLLDWIENEFIWVLSHIPGFA 306
LATLIGKAIIKTHIQT+FIISVCNNQLLDWIENEFIWVLSHIPGFA
Sbjct: 529 LATLIGKAIIKTHIQTIFIISVCNNQLLDWIENEFIWVLSHIPGFA 666
>TC206661
Length = 541
Score = 331 bits (848), Expect = 4e-91
Identities = 157/178 (88%), Positives = 170/178 (95%)
Frame = +3
Query: 42 MLFSVAVGTLGVVLMTLGCLHEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYL 101
MLFSVAVGTLG+VLMTLGCLHEKHLEE LEYFRFGLWWV LGVASSIGLGSGLHTFVLYL
Sbjct: 3 MLFSVAVGTLGIVLMTLGCLHEKHLEELLEYFRFGLWWVPLGVASSIGLGSGLHTFVLYL 182
Query: 102 GPHIALFTIKAMQCGRVDLKSAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSS 161
GPHIALFT+KA+QCGRVDLKSAPYDT QL+R SWLDKNCS+FGPPLFQS Y ++VPLSS
Sbjct: 183 GPHIALFTLKAVQCGRVDLKSAPYDTTQLSRSASWLDKNCSEFGPPLFQSVYDSQVPLSS 362
Query: 162 ILPQVQMEAVLWGLGTAIGELPPYFISRAARLSGSRMDAMEELDSEDKGVLNQIKCWF 219
ILPQVQ+EA+LWG+GTAIGELPPYFISRAARLSGSR+DAMEELDSEDK VL++IKCWF
Sbjct: 363 ILPQVQLEAILWGIGTAIGELPPYFISRAARLSGSRVDAMEELDSEDKRVLSRIKCWF 536
>TC206659 similar to UP|O23033 (O23033) YUP8H12.2 protein, partial (40%)
Length = 926
Score = 226 bits (576), Expect(3) = 5e-86
Identities = 111/146 (76%), Positives = 129/146 (88%), Gaps = 4/146 (2%)
Frame = +3
Query: 256 FWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIENEFIWVLSHIPGFASVLPKLTAN 315
F NFFLATLIGKAIIKTHIQT+FIISVCNNQLL WIENEFIWVLSHIPGFASVLP++T++
Sbjct: 180 FGNFFLATLIGKAIIKTHIQTIFIISVCNNQLLHWIENEFIWVLSHIPGFASVLPRVTSS 359
Query: 316 LHAMKDKYLKA-PHPVSPNTKGKKWDFSFTSIWNTVVWLMLMNFFIKIVNSTAQTHLKKQ 374
L AMKDKYLK HPVSPN +G+KWDFSFT +WNT VWLMLMNFF+KIVN+TAQ +LKKQ
Sbjct: 360 LRAMKDKYLKKDSHPVSPNKQGEKWDFSFTLVWNTGVWLMLMNFFVKIVNATAQRYLKKQ 539
Query: 375 QESEVAALTKKS---DSDTQ*NALII 397
QE++++ALT+KS DSD Q* ++
Sbjct: 540 QETQLSALTEKSTPTDSDAQ*IVYVL 617
Score = 102 bits (255), Expect(3) = 5e-86
Identities = 45/53 (84%), Positives = 48/53 (89%)
Frame = +2
Query: 212 LNQIKCWFFSHTQHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFWNFFLATL 264
L++IKCWF SH+QHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFW FF L
Sbjct: 47 LSRIKCWFLSHSQHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFWKFFSCNL 205
Score = 28.9 bits (63), Expect(3) = 5e-86
Identities = 15/20 (75%), Positives = 17/20 (85%), Gaps = 2/20 (10%)
Frame = +1
Query: 198 MDAMEELDSEDKGVL--NQI 215
+DAMEELDSEDK VL NQ+
Sbjct: 4 VDAMEELDSEDKRVLESNQV 63
>BG238027
Length = 292
Score = 158 bits (400), Expect = 4e-39
Identities = 77/97 (79%), Positives = 81/97 (83%)
Frame = +2
Query: 192 RLSGSRMDAMEELDSEDKGVLNQIKCWFFSHTQHLNFFTILVLASVPNPLFDLAGIMCGQ 251
RL G R AME ED L++IKCWF SH+QHLNFFTILVLASVPNPLFDLAGIMCGQ
Sbjct: 2 RLPGGRFFAMEN*YQEDTRFLSRIKCWFLSHSQHLNFFTILVLASVPNPLFDLAGIMCGQ 181
Query: 252 FGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLL 288
FGIPFW F LATLI KAIIKTHIQT+FIIS CNNQLL
Sbjct: 182 FGIPFWKFSLATLIAKAIIKTHIQTIFIISACNNQLL 292
>TC206662 similar to UP|O23033 (O23033) YUP8H12.2 protein, partial (9%)
Length = 455
Score = 44.3 bits (103), Expect(2) = 2e-09
Identities = 23/41 (56%), Positives = 33/41 (80%), Gaps = 3/41 (7%)
Frame = +3
Query: 361 KIVNSTAQTHLKKQQESEVAALTKKS---DSDTQ*NALIII 398
KIVN+T+Q +LKKQQE+++AALT+KS DSD Q* +++
Sbjct: 45 KIVNATSQRYLKKQQETQLAALTEKSTPTDSDAQ*IVYVLL 167
Score = 35.8 bits (81), Expect(2) = 2e-09
Identities = 13/16 (81%), Positives = 15/16 (93%)
Frame = +1
Query: 346 IWNTVVWLMLMNFFIK 361
+WNTVVWLMLMNFF +
Sbjct: 1 VWNTVVWLMLMNFFAR 48
>TC230760 weakly similar to UP|Q9LVG7 (Q9LVG7) Ankyrin-like protein, partial
(11%)
Length = 976
Score = 30.0 bits (66), Expect = 2.1
Identities = 13/27 (48%), Positives = 17/27 (62%)
Frame = +3
Query: 78 WWVALGVASSIGLGSGLHTFVLYLGPH 104
W L V +G+GS L +VLY+GPH
Sbjct: 780 WVSPLNVLKWVGVGSLLGYYVLYIGPH 860
>TC216916 similar to UP|Q9ARE3 (Q9ARE3) ZF-HD homeobox protein, partial (50%)
Length = 1604
Score = 30.0 bits (66), Expect = 2.1
Identities = 12/21 (57%), Positives = 16/21 (76%)
Frame = -2
Query: 73 FRFGLWWVALGVASSIGLGSG 93
FRFG WW+ +GV++S GSG
Sbjct: 379 FRFGFWWIRVGVSTS---GSG 326
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 29.3 bits (64), Expect = 3.6
Identities = 13/28 (46%), Positives = 16/28 (56%)
Frame = -3
Query: 135 SWLDKNCSQFGPPLFQSEYGTRVPLSSI 162
S+ K CS PPLF +G PLSS+
Sbjct: 908 SFCSKGCSDVQPPLFLDFFGVVKPLSSV 825
>TC207861 similar to UP|MPK7_ARATH (Q39027) Mitogen-activated protein kinase
homolog 7 (MAP kinase 7) (AtMPK7) , partial (67%)
Length = 911
Score = 28.5 bits (62), Expect = 6.2
Identities = 16/81 (19%), Positives = 37/81 (44%)
Frame = -3
Query: 295 FIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVWLM 354
F W+ +PG LP+++A +H ++ P +S + K + + ++ + W
Sbjct: 363 FSWLRHSVPGKIGFLPRISAKMHPTDQTSIEVP-*LSQHNKSSGARYQRVTTYSVINWPS 187
Query: 355 LMNFFIKIVNSTAQTHLKKQQ 375
+V ++ ++H+ K Q
Sbjct: 186 -----TPLVRASPKSHIFKSQ 139
>TC212580
Length = 388
Score = 28.1 bits (61), Expect = 8.1
Identities = 12/32 (37%), Positives = 20/32 (62%)
Frame = +3
Query: 393 NALIIIFAVHPARSKLKFKFQLFLKPPMEVTP 424
NA++ +F+VH R ++K+K + FL P P
Sbjct: 33 NAVLYLFSVHL*RGEVKYKLRYFLPYPCITCP 128
>TC225624 similar to UP|Q7Y240 (Q7Y240) Thioredoxin peroxidase 1, complete
Length = 834
Score = 28.1 bits (61), Expect = 8.1
Identities = 12/29 (41%), Positives = 17/29 (58%)
Frame = +3
Query: 13 FKTLKYSTLAVIQYIKKTMLYLLAKGGWL 41
F L + + +QY+ KT YLL K GW+
Sbjct: 3 FLVLPLNLINKLQYLTKTRDYLLYKSGWI 89
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.331 0.143 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,639,707
Number of Sequences: 63676
Number of extensions: 328609
Number of successful extensions: 2183
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 2175
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2181
length of query: 435
length of database: 12,639,632
effective HSP length: 100
effective length of query: 335
effective length of database: 6,272,032
effective search space: 2101130720
effective search space used: 2101130720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC145222.7


