

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.4 - phase: 0
(521 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
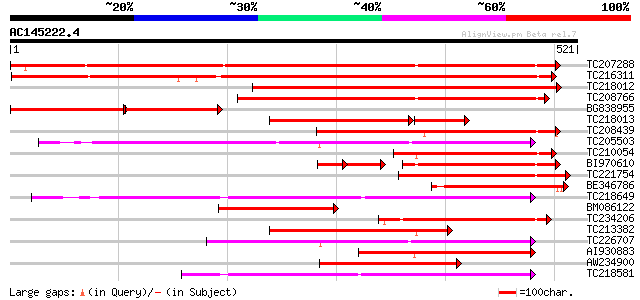
Score E
Sequences producing significant alignments: (bits) Value
TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete 534 e-152
TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete 517 e-147
TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporte... 498 e-141
TC208766 similar to UP|STA_RICCO (Q10710) Sugar carrier protein ... 352 2e-97
BG838955 similar to GP|5734440|emb hexose transporter {Lycopersi... 194 5e-90
TC218013 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporte... 193 3e-73
TC208439 weakly similar to UP|O04078 (O04078) Monosaccharid tran... 252 3e-67
TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete 205 5e-53
TC210054 homologue to UP|Q7XA52 (Q7XA52) Monosaccharide transpor... 194 6e-50
BI970610 163 3e-49
TC221754 similar to UP|Q06312 (Q06312) H(+)/MONOSACCHARIDE cotra... 178 6e-45
BE346786 similar to GP|5734440|emb hexose transporter {Lycopersi... 171 9e-43
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 170 1e-42
BM086122 similar to PIR|T07379|T07 hexose transport protein - to... 167 8e-42
TC234206 weakly similar to GB|AAO64833.1|29028908|BT005898 At1g3... 160 1e-39
TC213382 homologue to UP|Q7XA52 (Q7XA52) Monosaccharide transpor... 157 8e-39
TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 149 2e-36
AI930883 weakly similar to GP|4138724|emb hexose transporter {Vi... 145 3e-35
AW234900 weakly similar to GP|16945177|em STP5 protein {Arabidop... 138 7e-33
TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F2... 134 1e-31
>TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete
Length = 1813
Score = 534 bits (1375), Expect = e-152
Identities = 265/511 (51%), Positives = 364/511 (70%), Gaps = 5/511 (0%)
Frame = +1
Query: 1 MAGGGFATSGGGE----FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKF 56
MAGGG G G+ +E K T +C++ A GG +FGYD+GVSGGV SM FLK+F
Sbjct: 28 MAGGGLTNGGPGKRAHLYEHKFTAYFAFTCVVGALGGSLFGYDLGVSGGVPSMDDFLKEF 207
Query: 57 FPAVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIA 116
FP VYR+ + +++YCKYD+Q L LFTSSLY +AL TFFAS+ TR GR+ ++++
Sbjct: 208 FPKVYRRKQMHLH-ETDYCKYDDQVLTLFTSSLYFSALVMTFFASFLTRKKGRKASIIVG 384
Query: 117 GFFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQL 176
F+AG NAAA+N+AMLI+GR+LLG G+GF NQAVP++LSE+AP++ RGA+N LFQ
Sbjct: 385 ALSFLAGAILNAAAKNIAMLIIGRVLLGGGIGFGNQAVPLYLSEMAPAKNRGAVNQLFQF 564
Query: 177 NVTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLE 236
GIL ANLVNY T KI +GWR+SLGLAG+PA + VG I +TPNSL+E+GRL+
Sbjct: 565 TTCAGILIANLVNYFTEKIH-PYGWRISLGLAGLPAFAMLVGGICCAETPNSLVEQGRLD 741
Query: 237 EGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIM-AIGLEIFQQ 295
+ K VL++IRGT+N+E EF +L EAS A+ VK FR LLK + Q+I+ A+G+ FQQ
Sbjct: 742 KAKQVLQRIRGTENVEAEFEDLKEASEEAQAVKSPFRTLLKRKYRPQLIIGALGIPAFQQ 921
Query: 296 FTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAG 355
TG N+I+FYAPV+F ++GF +ASL+S+ IT +++T++S++ VDK GRR LEAG
Sbjct: 922 LTGNNSILFYAPVIFQSLGFGANASLFSSFITNGALLVATVISMFLVDKYGRRKFFLEAG 1101
Query: 356 VQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETF 415
+M I+ +L + H ++ KG + F+V+++ FV A+ SWGPLGWL+PSE F
Sbjct: 1102FEMICCMIITGAVLAVNF-GHGKEIGKGVSAFLVVVIFLFVLAYGRSWGPLGWLVPSELF 1278
Query: 416 PLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPET 475
PLE RS+ QS+ VCVNM+FT ++AQ FL LCH KFGIFL F+ ++ MS FV FL+PET
Sbjct: 1279PLEIRSSAQSIVVCVNMIFTALVAQLFLMSLCHLKFGIFLLFASLIIFMSFFVFFLLPET 1458
Query: 476 KNIPIEEMTERVWKQHWFWKRFMEDDNEKVS 506
K +PIEE+ +++ HWFW+RF+ D + + S
Sbjct: 1459KKVPIEEI-YLLFENHWFWRRFVTDQDPETS 1548
>TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete
Length = 1925
Score = 517 bits (1331), Expect = e-147
Identities = 278/509 (54%), Positives = 358/509 (69%), Gaps = 8/509 (1%)
Frame = +1
Query: 2 AGGGFATSGGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVY 61
A GG + GG E+ +TP V ++CI+AA GGL+FGYD+G+SGGVTSM PFL KFFP+V+
Sbjct: 103 AVGGISNGGGKEYPGSLTPFVTVTCIVAAMGGLIFGYDIGISGGVTSMDPFLLKFFPSVF 282
Query: 62 RKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFI 121
RK + ++ YC+YD+Q L +FTSSLYLAAL S+ AS TR GR+L+ML G F+
Sbjct: 283 RKKNSDKTVNQ-YCQYDSQTLTMFTSSLYLAALLSSLVASTVTRRFGRKLSMLFGGLLFL 459
Query: 122 AGVAFNAAAQNLAMLIVGRILLGCGVGFANQA---VPVFLSEIAPSRIRGAL--NILFQL 176
G N AQ++ MLIVGRILLG G+GFANQ +P+ I R G L N Q
Sbjct: 460 VGALINGFAQHVWMLIVGRILLGFGIGFANQVCATLPI*NGFIQI*RSIGTLAFNCQSQF 639
Query: 177 NVTIGILFANLVNYGTNKISGGWGWRLSLGL-AGIPALLLTVGAIVVVDTPNSLIERGRL 235
+ G L+ K WGW++ + A +PAL++TVG++V+ DTPNS+IERG
Sbjct: 640 GIPRGQCVELLLWL---KSMVAWGWKIEVWEGAMVPALIITVGSLVLPDTPNSMIERGDR 810
Query: 236 EEGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQ 295
E+ KA L++IRG DN++ EF +L AS + +V+ +RNLL+ + + MA+ + FQQ
Sbjct: 811 EKAKAQLQRIRGIDNVDEEFNDLVAASESSSQVEHPWRNLLQRKYRPHLTMAVLIPFFQQ 990
Query: 296 FTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAG 355
TGIN IMFYAPVLF+++GFK+DA+L SAVITG VNV++T VSIY VDK GRR L LE G
Sbjct: 991 LTGINVIMFYAPVLFSSIGFKDDAALMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGG 1170
Query: 356 VQMFLSQIVIAIILGIKV-TD-HSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSE 413
VQM + Q V+A +G K TD + DL K YAI VV+ +C +VSAFAWSWGPLGWL+PSE
Sbjct: 1171VQMLICQAVVAAAIGAKFGTDGNPGDLPKWYAIVVVLFICIYVSAFAWSWGPLGWLVPSE 1350
Query: 414 TFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVP 473
FPLE RSA QS+ V VNMLFTF+IAQ FL+MLCH KFG+FLFF+ +VLIM+ FV F +P
Sbjct: 1351IFPLEIRSAAQSINVSVNMLFTFLIAQVFLTMLCHMKFGLFLFFAFFVLIMTFFVYFFLP 1530
Query: 474 ETKNIPIEEMTERVWKQHWFWKRFMEDDN 502
ETK IPIEEM + VW+ H FW RF+E D+
Sbjct: 1531ETKGIPIEEMGQ-VWQAHPFWSRFVEHDD 1614
>TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporter 4, partial
(54%)
Length = 1142
Score = 498 bits (1282), Expect = e-141
Identities = 247/285 (86%), Positives = 268/285 (93%), Gaps = 1/285 (0%)
Frame = +3
Query: 224 DTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQ 283
DTPNSLIERGRLEEGK VLKKIRGTDNIE EF EL EASRVAKEVK FRNLLK Q
Sbjct: 3 DTPNSLIERGRLEEGKTVLKKIRGTDNIELEFQELVEASRVAKEVKHPFRNLLKRRNRPQ 182
Query: 284 VIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVD 343
++++I L+IFQQFTGINAIMFYAPVLFNT+GFKNDASLYSAVITGAVNVLST+VSIY VD
Sbjct: 183 LVISIALQIFQQFTGINAIMFYAPVLFNTLGFKNDASLYSAVITGAVNVLSTVVSIYSVD 362
Query: 344 KLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSW 403
KLGRRMLLLEAGVQMFLSQ+VIAIILGIKVTDHSDDLSKG AI VV++VCTFVS+FAWSW
Sbjct: 363 KLGRRMLLLEAGVQMFLSQVVIAIILGIKVTDHSDDLSKGIAILVVVMVCTFVSSFAWSW 542
Query: 404 GPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLI 463
GPLGWLIPSETFPLETRSAGQSVTVCVN+LFTFVIAQAFLSMLCHFKFGIFLFFSGWVL+
Sbjct: 543 GPLGWLIPSETFPLETRSAGQSVTVCVNLLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLV 722
Query: 464 MSIFVLFLVPETKNIPIEEMTERVWKQHWFWKRFMED-DNEKVSN 507
MS+FVLFL+PETKN+PIEEMTERVWKQHWFWKRF++D +EKV++
Sbjct: 723 MSVFVLFLLPETKNVPIEEMTERVWKQHWFWKRFIDDAADEKVAH 857
>TC208766 similar to UP|STA_RICCO (Q10710) Sugar carrier protein A, partial
(55%)
Length = 1202
Score = 352 bits (904), Expect = 2e-97
Identities = 168/287 (58%), Positives = 222/287 (76%)
Frame = +1
Query: 210 IPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVAKEVK 269
+PALL+TVG I + DTPNSLIERG E+G+ +L+KIRGT ++ EF ++ +AS +AK +K
Sbjct: 4 VPALLMTVGGIFLPDTPNSLIERGLAEKGRKLLEKIRGTKEVDAEFQDMVDASELAKSIK 183
Query: 270 QAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGA 329
FRN+L+ +++MAI + FQ TGIN+I+FYAPVLF ++GF DASL S+ +TG
Sbjct: 184 HPFRNILERRYRPELVMAIFMPTFQILTGINSILFYAPVLFQSMGFGGDASLISSALTGG 363
Query: 330 VNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVV 389
V ST +SI VD+LGRR+LL+ G+QM QI++AIILG+K +LSKG++I VV
Sbjct: 364 VLASSTFISIATVDRLGRRVLLVSGGLQMITCQIIVAIILGVKF-GADQELSKGFSILVV 540
Query: 390 ILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHF 449
+++C FV AF WSWGPLGW +PSE FPLE RSAGQ +TV VN+LFTF+IAQAFL++LC F
Sbjct: 541 VVICLFVVAFGWSWGPLGWTVPSEIFPLEIRSAGQGITVAVNLLFTFIIAQAFLALLCSF 720
Query: 450 KFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMTERVWKQHWFWKR 496
KFGIFLFF+GW+ IM+IFV +PETK IPIEEM+ +W++HWFWKR
Sbjct: 721 KFGIFLFFAGWITIMTIFVYLFLPETKGIPIEEMS-FMWRRHWFWKR 858
>BG838955 similar to GP|5734440|emb hexose transporter {Lycopersicon
esculentum}, partial (36%)
Length = 631
Score = 194 bits (492), Expect(2) = 5e-90
Identities = 97/110 (88%), Positives = 100/110 (90%), Gaps = 1/110 (0%)
Frame = -3
Query: 1 MAGGGFATSGG-GEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPA 59
MAGGGF + GG G+FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSM PFL KFFP
Sbjct: 593 MAGGGFTSGGGAGDFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMPPFLXKFFPT 414
Query: 60 VYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGR 109
VYRKTV E GLDSNYCKYDNQGLQLFTSSLYLA LTSTFFASYTTR +GR
Sbjct: 413 VYRKTVEEKGLDSNYCKYDNQGLQLFTSSLYLAGLTSTFFASYTTRRLGR 264
Score = 156 bits (394), Expect(2) = 5e-90
Identities = 80/89 (89%), Positives = 84/89 (93%)
Frame = -2
Query: 107 MGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRI 166
+G++LTMLIAG FFI GV NAAAQ+LAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRI
Sbjct: 273 VGKKLTMLIAGVFFICGVVLNAAAQDLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRI 94
Query: 167 RGALNILFQLNVTIGILFANLVNYGTNKI 195
R ALNILFQLNVTIGILFANLVNYGTNKI
Sbjct: 93 RRALNILFQLNVTIGILFANLVNYGTNKI 7
>TC218013 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporter 4, partial
(33%)
Length = 568
Score = 193 bits (491), Expect(2) = 3e-73
Identities = 102/133 (76%), Positives = 114/133 (85%)
Frame = +2
Query: 239 KAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTG 298
K LK+ NIE EF EL EASRVAKEVK FRNLLK Q+++++ L+IFQQFTG
Sbjct: 14 KQCLKRFGALYNIELEFQELLEASRVAKEVKHPFRNLLKRRNRPQLVISVALQIFQQFTG 193
Query: 299 INAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQM 358
INAIMFYAPVLFNT+GFKNDASLYSAVITGAVNVLST+VSIY VDK+GRR+LLLEAGVQM
Sbjct: 194 INAIMFYAPVLFNTLGFKNDASLYSAVITGAVNVLSTVVSIYSVDKVGRRILLLEAGVQM 373
Query: 359 FLSQIVIAIILGI 371
FLSQ+VIAIILGI
Sbjct: 374 FLSQVVIAIILGI 412
Score = 100 bits (250), Expect(2) = 3e-73
Identities = 45/50 (90%), Positives = 48/50 (96%)
Frame = +3
Query: 373 VTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSA 422
VTDHSDDLSKG AI VV++VCTFVS+FAWSWGPLGWLIPSETFPLETRSA
Sbjct: 417 VTDHSDDLSKGIAILVVVMVCTFVSSFAWSWGPLGWLIPSETFPLETRSA 566
>TC208439 weakly similar to UP|O04078 (O04078) Monosaccharid transport
protein, partial (43%)
Length = 936
Score = 252 bits (643), Expect = 3e-67
Identities = 126/230 (54%), Positives = 164/230 (70%), Gaps = 6/230 (2%)
Frame = +1
Query: 283 QVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFV 342
Q++ AI + FQQFTG+N I FYAP+LF T+GF + ASL SAVI G+ +ST+VSI V
Sbjct: 7 QLVFAICIPFFQQFTGLNVITFYAPILFRTIGFGSGASLMSAVIIGSFKPVSTLVSILLV 186
Query: 343 DKLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDHSDD--LSKGYAIFVVILVCTFVSAFA 400
DK GRR L LE G QM + QI++ I + + + + L K YAI VV ++C +VS FA
Sbjct: 187 DKFGRRTLFLEGGAQMLICQIIMTIAIAVTFGTNGNPGTLPKWYAIVVVGIICVYVSGFA 366
Query: 401 WSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGW 460
WSWGPLGWLIPSE FPLE R A QS+TV VNM+ TF IAQ F SMLCH KFG+F+FF +
Sbjct: 367 WSWGPLGWLIPSEIFPLEIRPAAQSITVGVNMISTFFIAQFFTSMLCHMKFGLFIFFGCF 546
Query: 461 VLIMSIFVLFLVPETKNIPIEEMTERVWKQHWFWKRFMEDD----NEKVS 506
V+IM+ F+ L+PETK IP+EEM+ VW++H W +F+E D N+K++
Sbjct: 547 VVIMTTFIYKLLPETKGIPLEEMS-MVWQKHPIWGKFLESDITTQNDKIN 693
>TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete
Length = 1928
Score = 205 bits (521), Expect = 5e-53
Identities = 136/478 (28%), Positives = 235/478 (48%), Gaps = 21/478 (4%)
Frame = +1
Query: 27 IMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDNQGLQLFT 86
++A+ ++ GYD+GV G A+Y K L K ++ +++
Sbjct: 271 MLASMTSILLGYDIGVMSGA------------AIYIKRDL---------KVSDEQIEILL 387
Query: 87 SSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCG 146
+ L +L + A T+ +GRR T+ + G F+ G + + L+ GR + G G
Sbjct: 388 GIINLYSLIGSCLAGRTSDWIGRRYTIGLGGAIFLVGSTLMGFYPHYSFLMCGRFVAGIG 567
Query: 147 VGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLG 206
+G+A PV+ +E++P+ RG L ++ + GIL + NYG +K++ GWR+ LG
Sbjct: 568 IGYALMIAPVYTAEVSPASSRGFLTSFPEVFINGGILLGYISNYGFSKLTLKVGWRMMLG 747
Query: 207 LAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVAK 266
+ IP+++LT G + + ++P L+ RGRL E + VL K +D+ E L L E + A
Sbjct: 748 VGAIPSVVLTEGVLAMPESPRWLVMRGRLGEARKVLNKT--SDSKEEAQLRLAEIKQAAG 921
Query: 267 EVKQAFRNLLKGEKGGQ-------------------VIMAIGLEIFQQFTGINAIMFYAP 307
+ ++++ K VI A+G+ FQQ +G++A++ Y+P
Sbjct: 922 IPESCNDDVVQVNKQSNGEGVWKELFLYPTPAIRHIVIAALGIHFFQQASGVDAVVLYSP 1101
Query: 308 VLFNTVGFKNDA-SLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIA 366
+F G ND L + V G V + + + + +D++GRR LLL + M LS + +A
Sbjct: 1102RIFEKAGITNDTHKLLATVAVGFVKTVFILAATFTLDRVGRRPLLLSSVGGMVLSLLTLA 1281
Query: 367 IILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSV 426
I + V DHS+ + +V +V+ F+ GP+ W+ SE FPL R+ G +
Sbjct: 1282I--SLTVIDHSERKLMWAVGSSIAMVLAYVATFSIGAGPITWVYSSEIFPLRLRAQGAAA 1455
Query: 427 TVCVNMLFTFVIAQAFLSMLCHFKF-GIFLFFSGWVLIMSIFVLFLVPETKNIPIEEM 483
V VN + V++ FLS+ G F + G + IF ++PET+ +E+M
Sbjct: 1456GVAVNRTTSAVVSMTFLSLTRAITIGGAFFLYCGIATVGWIFFYTVLPETRGKTLEDM 1629
>TC210054 homologue to UP|Q7XA52 (Q7XA52) Monosaccharide transporter, partial
(31%)
Length = 620
Score = 194 bits (494), Expect = 6e-50
Identities = 91/152 (59%), Positives = 115/152 (74%), Gaps = 2/152 (1%)
Frame = +2
Query: 353 EAGVQMFLSQIVIAIILGIK--VTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLI 410
E GVQM + Q V+A +G K + + DL K YA+ VV+ +C +VSAFAWSWGPLGWL+
Sbjct: 2 EGGVQMVICQAVVAAAIGAKFGIDGNPGDLPKWYAVVVVLFICIYVSAFAWSWGPLGWLV 181
Query: 411 PSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLF 470
PSE FPLE RSA QS+ V VNM FTF+IAQ FL+MLCH KFG+F+ F+ +VLIM+ F+ F
Sbjct: 182 PSEIFPLEIRSAAQSINVSVNMFFTFLIAQVFLTMLCHMKFGLFILFAFFVLIMTFFIYF 361
Query: 471 LVPETKNIPIEEMTERVWKQHWFWKRFMEDDN 502
+PETK IPIEEM + VWK H FW RF+E+D+
Sbjct: 362 FLPETKGIPIEEMNQ-VWKAHPFWSRFVENDD 454
>BI970610
Length = 742
Score = 163 bits (413), Expect(3) = 3e-49
Identities = 73/145 (50%), Positives = 103/145 (70%)
Frame = +1
Query: 362 QIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRS 421
QI+ +L + H ++ KG + F+V+++ FV A+ SWGPLGWL+PSE FPLE RS
Sbjct: 208 QIITGAVLAVNF-GHGKEIGKGVSAFLVVVIFLFVLAYGRSWGPLGWLVPSELFPLEIRS 384
Query: 422 AGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIE 481
+ QS+ VCVNM+FT ++AQ FL LCH KFGIFL F+ ++ MS FV FL+PETK +PIE
Sbjct: 385 SAQSIVVCVNMIFTALVAQLFLMSLCHLKFGIFLLFASLIIFMSFFVFFLLPETKKVPIE 564
Query: 482 EMTERVWKQHWFWKRFMEDDNEKVS 506
E+ +++ HWFW+RF+ D + + S
Sbjct: 565 EI-YLLFENHWFWRRFVTDQDPETS 636
Score = 37.7 bits (86), Expect(3) = 3e-49
Identities = 16/39 (41%), Positives = 27/39 (69%)
Frame = +2
Query: 307 PVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKL 345
P F + GF +ASL+S+ IT +++T++S++ VDKL
Sbjct: 98 PCHFQSXGFXXNASLFSSXITNGALLVATVISMFLVDKL 214
Score = 33.5 bits (75), Expect(3) = 3e-49
Identities = 14/27 (51%), Positives = 20/27 (73%)
Frame = +3
Query: 284 VIMAIGLEIFQQFTGINAIMFYAPVLF 310
+I A+G+ FQQ N+I+FYAPV+F
Sbjct: 30 MIGALGIXAFQQLXXNNSILFYAPVIF 110
>TC221754 similar to UP|Q06312 (Q06312) H(+)/MONOSACCHARIDE cotransporter,
partial (24%)
Length = 582
Score = 178 bits (451), Expect = 6e-45
Identities = 85/160 (53%), Positives = 109/160 (68%), Gaps = 2/160 (1%)
Frame = +3
Query: 358 MFLSQIVIAIILGIKV-TDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFP 416
M + QI +++ +K S G A ++ +C FV+AFAWSWGPLGWL+PSE P
Sbjct: 3 MLICQIATGVMIAMKFGVSGEGSFSSGEANLILFFICAFVAAFAWSWGPLGWLVPSEICP 182
Query: 417 LETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETK 476
LE RSAGQ++ V VNMLFTF IAQ FL MLCH KFG+F FF+ +VLIM+IF+ L+PETK
Sbjct: 183 LEVRSAGQAINVAVNMLFTFAIAQVFLVMLCHLKFGLFFFFAAFVLIMTIFIAMLLPETK 362
Query: 477 NIPIEEMTERVWKQHWFWKRFM-EDDNEKVSNADYPKIKN 515
NIPIEEM VW+ HWFW + + D+++ A IKN
Sbjct: 363 NIPIEEM-HTVWRSHWFWSKIVPHADDDRKPEAAQS*IKN 479
>BE346786 similar to GP|5734440|emb hexose transporter {Lycopersicon
esculentum}, partial (19%)
Length = 435
Score = 171 bits (432), Expect = 9e-43
Identities = 84/132 (63%), Positives = 97/132 (72%), Gaps = 6/132 (4%)
Frame = -1
Query: 388 VVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLC 447
VV++VC GPLGW IPSE FPLETRS GQ ++VCVN LFTFVI QA SMLC
Sbjct: 435 VVVMVC-----ICMVMGPLGWFIPSEIFPLETRSVGQGLSVCVNFLFTFVIGQAVFSMLC 271
Query: 448 HFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMTERVWKQHWFWKRFMEDDN----E 503
F+FGIF FF GW+LIMS FVLFL PET N+PIEEM ERVWKQHW WKRF+++D+ E
Sbjct: 270 LFRFGIFFFFYGWILIMSTFVLFLFPETNNVPIEEMAERVWKQHWLWKRFIDEDDCVKEE 91
Query: 504 KV--SNADYPKI 513
KV N + P+I
Sbjct: 90 KVVTGN*ECPRI 55
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 170 bits (431), Expect = 1e-42
Identities = 130/468 (27%), Positives = 222/468 (46%), Gaps = 5/468 (1%)
Frame = +2
Query: 21 IVIISCIM-AATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDN 79
I + +C++ A G + FG+ G + S + + GL +
Sbjct: 167 ISVFACVLIVALGPIQFGFTAGYTSPTQSA--------------IINDLGLSVSE----- 289
Query: 80 QGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVG 139
LF S + A+ + +GR+ +++IA I G + A++ + L +G
Sbjct: 290 --FSLFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIASIPNIIGWLAISFAKDSSFLYMG 463
Query: 140 RILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGW 199
R+L G GVG + VPV+++EI+P +RG L + QL+VTIGI+ A L+
Sbjct: 464 RLLEGFGVGIISYTVPVYIAEISPPNLRGGLVSVNQLSVTIGIMLAYLLGIFVE------ 625
Query: 200 GWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTD-NIEPEFLEL 258
WR+ + +P +L + ++P L + G EE + L+ +RG D +I E E+
Sbjct: 626 -WRILAIIGILPCTILIPALFFIPESPRWLAKMGMTEEFETSLQVLRGFDTDISVEVNEI 802
Query: 259 CEA-SRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGF-K 316
A + + F +L + +++ IGL I QQ +GIN ++FY+ +F G
Sbjct: 803 KRAVASTNTRITVRFADLKQRRYWLPLMIGIGLLILQQLSGINGVLFYSSTIFRNAGISS 982
Query: 317 NDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKVT-D 375
+DA+ + GAV VL+T ++++ DK GRR+LL+ + M S +V+AI IK +
Sbjct: 983 SDAATFG---VGAVQVLATSLTLWLADKSGRRLLLIVSATGMSFSLLVVAITFYIKASIS 1153
Query: 376 HSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFT 435
+ L + ++ V V AF+ G + W+I SE P+ + SV N LF+
Sbjct: 1154ETSSLYGILSTLSLVGVVAMVIAFSLGMGAMPWIIMSEILPINIKGLAGSVATLANWLFS 1333
Query: 436 FVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEM 483
+++ +L G F ++ + +FV VPETK IEE+
Sbjct: 1334WLVTLTANMLLDWSSGGTFTIYAVVCALTVVFVTIWVPETKGKTIEEI 1477
>BM086122 similar to PIR|T07379|T07 hexose transport protein - tomato,
partial (20%)
Length = 428
Score = 167 bits (424), Expect = 8e-42
Identities = 80/110 (72%), Positives = 95/110 (85%)
Frame = +2
Query: 193 NKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIE 252
++I GGWGWRLSLGL G+PALLLT+GA +VVDTPNSLIERG LEEGK+VL+KIRG DNIE
Sbjct: 92 DRIKGGWGWRLSLGLGGLPALLLTLGAFLVVDTPNSLIERGHLEEGKSVLRKIRGIDNIE 271
Query: 253 PEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAI 302
PEFLEL +ASRVAKEVK FRN+LK Q++++I L++FQQFTGIN I
Sbjct: 272 PEFLELLDASRVAKEVKHPFRNILKRRNRSQLVISIALQVFQQFTGINVI 421
>TC234206 weakly similar to GB|AAO64833.1|29028908|BT005898 At1g34580
{Arabidopsis thaliana;} , partial (23%)
Length = 740
Score = 160 bits (405), Expect = 1e-39
Identities = 73/162 (45%), Positives = 113/162 (69%), Gaps = 3/162 (1%)
Frame = +1
Query: 340 YFVD--KLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDH-SDDLSKGYAIFVVILVCTFV 396
YF++ K+ ++L++ V M +I ++ +L + H + D+SKG A+ V++L+C +
Sbjct: 94 YFIN*FKISVKLLIIIICVGML--KIAVSALLAMVTGVHGTKDISKGNAMLVLVLLCFYD 267
Query: 397 SAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLF 456
+ F WSWGPL WLIPSE FPL+ R+ GQS+ V V + F ++Q FL+MLCHFKFG FLF
Sbjct: 268 AGFGWSWGPLTWLIPSEIFPLKIRTTGQSIAVGVQFIALFALSQTFLTMLCHFKFGAFLF 447
Query: 457 FSGWVLIMSIFVLFLVPETKNIPIEEMTERVWKQHWFWKRFM 498
++ W+ +M++F++F +PETK IP+E M +W +HWFW RF+
Sbjct: 448 YTVWIAVMTLFIMFFLPETKGIPLESM-YTIWGKHWFWGRFV 570
>TC213382 homologue to UP|Q7XA52 (Q7XA52) Monosaccharide transporter, partial
(35%)
Length = 624
Score = 157 bits (398), Expect = 8e-39
Identities = 84/172 (48%), Positives = 118/172 (67%), Gaps = 3/172 (1%)
Frame = +1
Query: 239 KAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTG 298
KA L+++RG D++E EF +L AS +++V+ +RNLL+ + + MA+ + FQQ TG
Sbjct: 1 KAQLRRVRGIDDVEEEFNDLVAASESSRKVEHPWRNLLQRKYRPHLTMAVLIPFFQQLTG 180
Query: 299 INAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQM 358
IN IMFYAPVLF+++GFK+D++L SAVITG VNV++T VSIY VDK GRR L LE GVQM
Sbjct: 181 INVIMFYAPVLFSSIGFKDDSALMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGVQM 360
Query: 359 FLSQIVIAIILGIK---VTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLG 407
+ Q V+A +G K + + DL+K YA VV+ +C +V + G LG
Sbjct: 361 VICQAVVAAAIGAKFGNLMETLGDLAKWYAXVVVLFICIYVFSIWLVMGSLG 516
>TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 2338
Score = 149 bits (377), Expect = 2e-36
Identities = 98/321 (30%), Positives = 162/321 (49%), Gaps = 19/321 (5%)
Frame = +3
Query: 182 ILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAV 241
IL + NYG +K++ GWRL LG+ IP++L+ V + + ++P L+ +GRL E K V
Sbjct: 3 ILIGYISNYGFSKLALRLGWRLMLGVGAIPSILIGVAVLAMPESPRWLVAKGRLGEAKRV 182
Query: 242 LKKI-RGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQV---------------- 284
L KI + ++ + + + ++ + K G V
Sbjct: 183 LYKISESEEEARLRLADIKDTAGIPQDCDDDVVLVSKQTHGHGVWRELFLHPTPAVRHIF 362
Query: 285 IMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDA-SLYSAVITGAVNVLSTIVSIYFVD 343
I ++G+ F Q TGI+A++ Y+P +F G K+D L + V G V +S +V+ +F+D
Sbjct: 363 IASLGIHFFAQATGIDAVVLYSPRIFEKAGIKSDNYRLLATVAVGFVKTVSILVATFFLD 542
Query: 344 KLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSW 403
+ GRR+LLL + + LS + + L + V DHS + V ++V+ F+
Sbjct: 543 RAGRRVLLLCSVSGLILSLLTLG--LSLTVVDHSQTTLNWAVGLSIAAVLSYVATFSIGS 716
Query: 404 GPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKF-GIFLFFSGWVL 462
GP+ W+ SE FPL R+ G ++ VN + + VIA FLS+ G F F+G
Sbjct: 717 GPITWVYSSEIFPLRLRAQGVAIGAAVNRVTSGVIAMTFLSLQKAITIGGAFFLFAGVAA 896
Query: 463 IMSIFVLFLVPETKNIPIEEM 483
+ IF L+PET+ +EE+
Sbjct: 897 VAWIFHYTLLPETRGKTLEEI 959
>AI930883 weakly similar to GP|4138724|emb hexose transporter {Vitis
vinifera}, partial (31%)
Length = 505
Score = 145 bits (367), Expect = 3e-35
Identities = 82/165 (49%), Positives = 105/165 (62%), Gaps = 2/165 (1%)
Frame = +2
Query: 321 LYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILG--IKVTDHSD 378
L SAVITG VN +T VSIY VDK RR L LE GVQM + Q V+ +G + +
Sbjct: 2 LMSAVITGVVNADATCVSIYSVDKWCRRALFLEGGVQMLICQAVVVTAIGA*LGTDGNPC 181
Query: 379 DLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVI 438
DL + AI V + + +VSAFAW W L WL+PSE F E RSA QS+TV VNML+TF+I
Sbjct: 182 DLPQWNAIVVALYIGIYVSAFAW*WRTLCWLVPSEIFHFEIRSAAQSITVSVNMLYTFLI 361
Query: 439 AQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEM 483
A FL+MLCH + LF++ ++LIM+ FV F +T I IE +
Sbjct: 362 AHVFLTMLCHMTDRMDLFYAFFLLIMTFFVYFF*LDTHGISIEHI 496
>AW234900 weakly similar to GP|16945177|em STP5 protein {Arabidopsis
thaliana}, partial (24%)
Length = 405
Score = 138 bits (347), Expect = 7e-33
Identities = 65/132 (49%), Positives = 91/132 (68%), Gaps = 1/132 (0%)
Frame = +2
Query: 285 IMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDK 344
+MAI + FQQ TGIN + FYAP +F +VG +DA+L SA+I GAVN++S +VS VD+
Sbjct: 8 VMAIAIPFFQQMTGINIVAFYAPNIFQSVGLGHDAALLSAIILGAVNLVSLLVSTAIVDR 187
Query: 345 LGRRMLLLEAGVQMFLSQIVIAIILGIKVTDH-SDDLSKGYAIFVVILVCTFVSAFAWSW 403
GRR L + G+ M + QI ++I+L + H + D+S G AI V++L+C + + F WSW
Sbjct: 188 FGRRFLFVTGGICMLVCQIAVSILLAVVTGVHGTKDMSNGSAIVVLVLLCCYTAGFGWSW 367
Query: 404 GPLGWLIPSETF 415
GPL WLIPSE F
Sbjct: 368 GPLTWLIPSEIF 403
>TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;} , partial (66%)
Length = 1340
Score = 134 bits (337), Expect = 1e-31
Identities = 94/328 (28%), Positives = 161/328 (48%), Gaps = 3/328 (0%)
Frame = +2
Query: 159 SEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVG 218
+EIAP+ + G L + QL++TIGI+ A L+ N WR+ L +P +L G
Sbjct: 2 TEIAPNHLIGGLGSVNQLSITIGIMLAYLLGLFVN-------WRILAILGILPCTVLIPG 160
Query: 219 AIVVVDTPNSLIERGRLEEGKAVLKKIRGTD-NIEPEFLELCEA-SRVAKEVKQAFRNLL 276
+ ++P L + G ++E + L+ +RG D +I E E+ + + K F +L
Sbjct: 161 LFFIPESPRWLAKMGMIDEFETSLQVLRGFDTDISVEVHEIKRSVASTGKRAAIRFADLK 340
Query: 277 KGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTI 336
+ +++ IGL + QQ +GIN I+FY+ +F G + + + V GAV V++T
Sbjct: 341 RKRYWFPLMVGIGLLVLQQLSGINGILFYSTTIFANAGISSSEA--ATVGLGAVQVIATG 514
Query: 337 VSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIK-VTDHSDDLSKGYAIFVVILVCTF 395
+S + VDK GRR+LL+ + M +S ++++I ++ V L I ++ +
Sbjct: 515 ISTWLVDKSGRRLLLIISSSVMTVSLLIVSIAFYLEGVVSEDSHLFSILGIVSIVGLVAM 694
Query: 396 VSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFL 455
V F+ GP+ WLI SE P+ + S+ N L ++ I +L G F
Sbjct: 695 VIGFSLGLGPIPWLIMSEILPVNIKGLAGSIATMGNWLISWGITMTANLLLNWSSGGTFT 874
Query: 456 FFSGWVLIMSIFVLFLVPETKNIPIEEM 483
++ F+ VPETK +EE+
Sbjct: 875 IYTVVAAFTIAFIAMWVPETKGRTLEEI 958
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,780,527
Number of Sequences: 63676
Number of extensions: 299655
Number of successful extensions: 2719
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 2614
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2646
length of query: 521
length of database: 12,639,632
effective HSP length: 102
effective length of query: 419
effective length of database: 6,144,680
effective search space: 2574620920
effective search space used: 2574620920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC145222.4


