

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.11 + phase: 0 /pseudo
(394 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
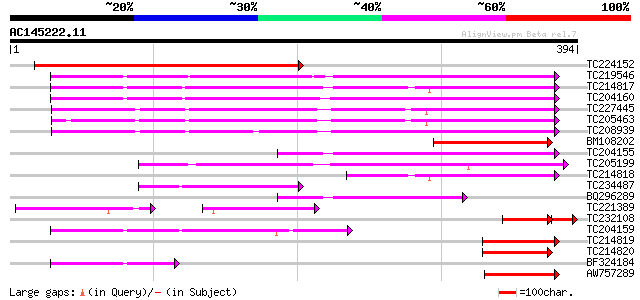
Score E
Sequences producing significant alignments: (bits) Value
TC224152 similar to UP|Q8L8N8 (Q8L8N8) 70kD heat shock protein, ... 326 1e-89
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 183 1e-46
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 181 5e-46
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 181 7e-46
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 159 3e-39
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 149 2e-36
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 140 1e-33
BM108202 weakly similar to GP|21618151|gb 70kD heat shock protei... 130 8e-31
TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, ... 117 1e-26
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 112 3e-25
TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous hea... 88 8e-18
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 76 2e-14
BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cyto... 72 3e-13
TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa p... 51 3e-13
TC232108 similar to UP|Q9SKY8 (Q9SKY8) 70kD heat shock protein (... 66 6e-12
TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 62 4e-10
TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat sh... 54 9e-08
TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight h... 54 2e-07
BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chapero... 49 4e-06
AW757289 42 5e-04
>TC224152 similar to UP|Q8L8N8 (Q8L8N8) 70kD heat shock protein, partial
(33%)
Length = 563
Score = 326 bits (836), Expect = 1e-89
Identities = 165/187 (88%), Positives = 172/187 (91%)
Frame = +1
Query: 18 GEEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVT 77
GEEKSS PEIAIGIDIGTSQCSVAVWNGS+VELLKN RNQK+MKS+VTFKD PSGGV+
Sbjct: 1 GEEKSSTFPEIAIGIDIGTSQCSVAVWNGSQVELLKNTRNQKIMKSYVTFKDNIPSGGVS 180
Query: 78 SQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHASKNLPFLVQTLDIGVRPFIAALVNNVW 137
SQ SHE EML G TIFNMKRLIGRVDTDPVVHA KNLPFLVQTLDIGVRPFIAALVNN+W
Sbjct: 181 SQLSHEDEMLSGATIFNMKRLIGRVDTDPVVHACKNLPFLVQTLDIGVRPFIAALVNNMW 360
Query: 138 RSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLR 197
RSTTPEEVLA+FLVELR M E LKR IRNVVLTVPVSFSRFQLTR+ERACAMAGLHVLR
Sbjct: 361 RSTTPEEVLAIFLVELRAMAEAQLKRRIRNVVLTVPVSFSRFQLTRIERACAMAGLHVLR 540
Query: 198 LMPEPTA 204
LMPEPTA
Sbjct: 541 LMPEPTA 561
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 183 bits (465), Expect = 1e-46
Identities = 118/357 (33%), Positives = 195/357 (54%), Gaps = 3/357 (0%)
Frame = +1
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 22 AIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 195
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V L PF V +P I + + EE+ +
Sbjct: 196 QNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCD-KPMIVVNYKGEEKKFSAEEISS 372
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M LV++R + E L ++N V+TVP F+ Q + A A++GL+VLR++ EPTA A+
Sbjct: 373 MVLVKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAI 552
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS++ E+ LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 553 AYG-LDKKASRK-----GEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGEDF 714
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 715 DNRMVNHFVSEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDF 894
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ +++V+LVGG + IP+ ++L
Sbjct: 895 YATITRARFEEMNMDLFRKCMEPVEKCLRDAKIDKSQVHEVVLVGGSTRIPKVHQLL 1065
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 181 bits (459), Expect = 5e-46
Identities = 120/361 (33%), Positives = 195/361 (53%), Gaps = 7/361 (1%)
Frame = +2
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 110 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAK--NQVAMNP 283
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + L PF V +P I + EE+ +
Sbjct: 284 INTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIP-GAADKPMIVVNYKGEEKQFAAEEISS 460
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L ++N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 461 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 640
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 641 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 802
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 803 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYE 970
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ V RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 971 GIDFYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQL 1150
Query: 382 L 382
L
Sbjct: 1151L 1153
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 181 bits (458), Expect = 7e-46
Identities = 115/357 (32%), Positives = 192/357 (53%), Gaps = 3/357 (0%)
Frame = +2
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 119 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 292
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V L PF V +P I + + EE+ +
Sbjct: 293 TNTVFDAKRLIGRRFSDASVQGDMKLWPFKVIP-GPAEKPMIVVNYKGEEKQFSAEEISS 469
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L++++ + E +L I+N V+TVP F+ Q + A ++GL+V+R++ EPTA A+
Sbjct: 470 MVLMKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAI 649
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 650 AYGLDKKAT------SSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 811
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 812 DNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDF 991
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 992 YTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLL 1162
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 159 bits (401), Expect = 3e-39
Identities = 112/361 (31%), Positives = 193/361 (53%), Gaps = 8/361 (2%)
Frame = +2
Query: 30 IGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKD-EAPSGGVTSQFSHEHEMLF 88
IGID+GT+ V V+ VE++ N + ++ S+V F D E G + +
Sbjct: 218 IGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVAFTDSERLIGEAAKNLAAVNPER- 394
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNN-VWRSTTPEEVL 146
TIF++KRLIGR D V L P+ + D +P+I + + + +PEE+
Sbjct: 395 --TIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKD--GKPYIQVKIKDGETKVFSPEEIS 562
Query: 147 AMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVA 206
AM L +++ E L + I + V+TVP F+ Q + A +AGL+V R++ EPTA A
Sbjct: 563 AMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAA 742
Query: 207 LLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGED 265
+ YG ++ G EK L+F++G G DV++ GV ++ A G T +GGED
Sbjct: 743 IAYGLDKK---------GGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGED 895
Query: 266 LLQNMMRHLLPDSENIFKRHGVE---EIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
Q +M + + + I K+HG + + +++ LR + A LS+Q V+V+++ L +
Sbjct: 896 FDQRIMEYFI---KLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFD 1066
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + + RA FEE+N ++F K + + + DA ++ I++++LVGG + IP+ ++
Sbjct: 1067GVDFSEPLTRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQL 1246
Query: 382 L 382
L
Sbjct: 1247L 1249
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 149 bits (377), Expect = 2e-36
Identities = 110/360 (30%), Positives = 190/360 (52%), Gaps = 7/360 (1%)
Frame = +3
Query: 30 IGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFG 89
IGID+ T C V VE++ N + ++ S+V F D G ++ +
Sbjct: 201 IGIDLTTYSC---VGVTDAVEIIANDQGNRITPSWVAFTDSERLIGEAAKIVAAVNPV-- 365
Query: 90 DTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNN-VWRSTTPEEVLA 147
TIF++KRLIGR D V L P+ + D +P+I + + + +PEE+ A
Sbjct: 366 RTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKD--GKPYIQVKIKDGETKVFSPEEISA 539
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ E L + I + V+TVP F+ Q + A +AGL+V R++ EPTA A+
Sbjct: 540 MILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 719
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G EK L+F++G G DV++ GV ++ A G T +GGED
Sbjct: 720 AYGLDKK---------GGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDF 872
Query: 267 LQNMMRHLLPDSENIFKRHGVE---EIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNS 322
Q +M + + + I K+HG + + +++ LR + A LS+Q V+V+++ L +
Sbjct: 873 DQRIMEYFI---KLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDG 1043
Query: 323 LKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ + + RA FEE+N ++F K + + + DA ++ I++++LVGG + IP+ ++L
Sbjct: 1044VDFSEPLTRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLL 1223
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 140 bits (353), Expect = 1e-33
Identities = 102/357 (28%), Positives = 182/357 (50%), Gaps = 4/357 (1%)
Frame = +3
Query: 30 IGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFG 89
IGID+GT+ V V+ VE++ N + ++ S+ E G + +
Sbjct: 180 IGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWSFTDSERLIGEAAKNLAAVNPER-- 353
Query: 90 DTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNN-VWRSTTPEEVLA 147
IF++KRLIGR D V L P+ + D +P+I + + + +PEE+ A
Sbjct: 354 -VIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKD--GKPYIQEKIKDGETKVFSPEEISA 524
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ E L + I + V F+ Q + A +AGL+V R++ EPTA A+
Sbjct: 525 MILTKMKETAEAFLGKKINDAV----AYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 692
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G EK L+F++G G DV++ GV ++ A G T +GGED
Sbjct: 693 AYGLDKK---------GGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDF 845
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
Q +M + + K+ ++ ++++ LR + A LS+Q V+V+++ L + +
Sbjct: 846 DQRIMEYFIKLINKKHKKDISKDSRALSKLRREAERAKRALSSQHQVRVEIESLFDGVDF 1025
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ + RA FEE+N ++F K + + + DA ++ I++++LVGG + IP+ ++L
Sbjct: 1026SEPLTRARFEELNNDLFRKTMGPVKKAMEDAGLQKNQIDEIVLVGGSTRIPKVQQLL 1196
>BM108202 weakly similar to GP|21618151|gb 70kD heat shock protein
{Arabidopsis thaliana}, partial (18%)
Length = 408
Score = 130 bits (328), Expect = 8e-31
Identities = 64/83 (77%), Positives = 73/83 (87%)
Frame = +3
Query: 295 LLRVATQDAIHQLSTQSSVQVDVDLGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLH 354
LLRVATQDAI QLS+Q+ VQVDVDLG+ LKICK V+R EFEEVN++VF KCESLI QCL
Sbjct: 6 LLRVATQDAIRQLSSQTIVQVDVDLGDGLKICKAVNREEFEEVNRKVFXKCESLIXQCLQ 185
Query: 355 DAKVEVGDINDVILVGGCSYIPQ 377
DAKVEV +NDVI+VGGCSYIP+
Sbjct: 186 DAKVEVEXVNDVIIVGGCSYIPR 254
>TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, partial
(75%)
Length = 1640
Score = 117 bits (292), Expect = 1e-26
Identities = 66/198 (33%), Positives = 113/198 (56%), Gaps = 2/198 (1%)
Frame = +2
Query: 187 ACAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTAT 246
A ++GL+V+R++ EPTA A+ YG ++ S EK LIF++G G DV++
Sbjct: 8 AGVISGLNVMRIINEPTAAAIAYGLDKKATSS------GEKNVLIFDLGGGTFDVSLLTI 169
Query: 247 AGGVSQIKALAGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIH 305
G+ ++KA AG T +GGED M+ H + + + K+ +++ LR A + A
Sbjct: 170 EEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKR 349
Query: 306 QLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDIN 364
LS+ + +++D L + + RA FEE+N ++F KC + +CL DAK++ ++
Sbjct: 350 TLSSTAQTTIEIDSLYEGIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVH 529
Query: 365 DVILVGGCSYIPQGWRIL 382
DV+LVGG + IP+ ++L
Sbjct: 530 DVVLVGGSTRIPKVQQLL 583
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 112 bits (280), Expect = 3e-25
Identities = 81/306 (26%), Positives = 151/306 (48%), Gaps = 7/306 (2%)
Frame = +3
Query: 90 DTIFNMKRLIGR-VDTDPVVHASKNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAM 148
+T+F KRLIGR D K +P+ + G A V + +P +V A
Sbjct: 303 NTLFGTKRLIGRRFDDSQTQKEMKMVPYKIVKAPNG-----DAWVEANGQQYSPSQVGAF 467
Query: 149 FLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALL 208
L +++ E++L + + V+TVP F+ Q + A +AGL V R++ EPTA AL
Sbjct: 468 VLTKMKETAESYLGKSVSKAVITVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALS 647
Query: 209 YGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLL 267
YG + E + +F++G G DV++ + GV ++KA G T +GGED
Sbjct: 648 YGM-----------NNKEGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFD 794
Query: 268 QNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDV-----DLGNS 322
++ L+ + + ++ ++ LR A + A +LS+ S ++++ D +
Sbjct: 795 NALLDFLVNEFKRTENIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGA 974
Query: 323 LKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ + R++FE + + E+ ++ CL DA V + ++++V+LVGG + +P+ ++
Sbjct: 975 KHLNITLTRSKFEALVNHLIERTKAPCKSCLKDANVSIKEVDEVLLVGGMTRVPKVQEVV 1154
Query: 383 LPTYAK 388
+ K
Sbjct: 1155SAIFGK 1172
>TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (68%)
Length = 1554
Score = 87.8 bits (216), Expect = 8e-18
Identities = 51/154 (33%), Positives = 87/154 (56%), Gaps = 6/154 (3%)
Frame = +2
Query: 235 GAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEI--- 290
G G DV++ G+ ++KA AG T +GGED M+ H + + FKR ++I
Sbjct: 8 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQE----FKRKNKKDISGN 175
Query: 291 -KSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESL 348
+++ LR A + A LS+ + +++D L + + RA FEE+N ++F KC
Sbjct: 176 PRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYSTITRARFEELNMDLFRKCMEP 355
Query: 349 IIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 356 VEKCLRDAKMDKRTVHDVVLVGGSTRIPKVQQLL 457
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 76.3 bits (186), Expect = 2e-14
Identities = 46/116 (39%), Positives = 69/116 (58%), Gaps = 1/116 (0%)
Frame = +1
Query: 90 DTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAM 148
+T+F+ KRLIGR +DPV+ K L PF V I +P I+ + EEV +M
Sbjct: 109 NTVFDAKRLIGRKFSDPVIQKDKMLWPFKV-VAGINDKPMISLNYKGQEKHLLAEEVSSM 285
Query: 149 FLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTA 204
L+++R + E +L+ P+ N V+TVP F+ Q A A+AGL+V+R++ EPTA
Sbjct: 286 VLIKMREIAEAYLETPVENAVVTVPAYFNDSQRKATIDAGAIAGLNVMRIINEPTA 453
>BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cytosolic
[imported] - spinach, partial (21%)
Length = 421
Score = 72.4 bits (176), Expect = 3e-13
Identities = 43/133 (32%), Positives = 73/133 (54%), Gaps = 1/133 (0%)
Frame = +1
Query: 187 ACAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTAT 246
A +AGL+V+R++ EPTA A+ YG ++ S EK LIF++G G DV++
Sbjct: 13 AGVIAGLNVMRIINEPTAAAIAYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTI 174
Query: 247 AGGVSQIKALAGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIH 305
G+ ++KA AG T +GGED M+ H + + + K+ +++ LR A + A
Sbjct: 175 EEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKHKKDINGNPRALRRLRTACERAKR 354
Query: 306 QLSTQSSVQVDVD 318
LS+ + +++D
Sbjct: 355 TLSSTAQTTIEID 393
>TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (36%)
Length = 827
Score = 50.8 bits (120), Expect(2) = 3e-13
Identities = 29/86 (33%), Positives = 46/86 (52%), Gaps = 5/86 (5%)
Frame = +1
Query: 135 NVWRST-----TPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACA 189
N W T +P ++ A L +++ E +L + I V+TVP F+ Q + A
Sbjct: 541 NAWVETNGQQYSPSQIGAFVLTKMKETAEAYLGKSISKAVITVPAYFNDAQRQATKDAGR 720
Query: 190 MAGLHVLRLMPEPTAVALLYGQQQQK 215
+AGL V R++ EPTA AL YG +++
Sbjct: 721 IAGLDVQRIINEPTAAALSYGMNKKE 798
Score = 41.6 bits (96), Expect(2) = 3e-13
Identities = 30/99 (30%), Positives = 46/99 (46%), Gaps = 2/99 (2%)
Frame = +3
Query: 5 EPAYTVASDSETTGEEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSF 64
+PAY + S + S P IGID+GT+ V+V G ++++N + S
Sbjct: 180 KPAYVAHNWSSLSRPFSSRPAGNDVIGIDLGTTNSCVSVMEGKNPKVIENSEGARTTPSV 359
Query: 65 VTF--KDEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGR 101
V F K E G + + + +T+F KRLIGR
Sbjct: 360 VAFNQKGELLVGTPXKRQAVTNPT---NTLFGTKRLIGR 467
>TC232108 similar to UP|Q9SKY8 (Q9SKY8) 70kD heat shock protein
(At2g32120/F22D22.13), partial (37%)
Length = 775
Score = 65.9 bits (159), Expect(2) = 6e-12
Identities = 29/35 (82%), Positives = 33/35 (93%)
Frame = +1
Query: 343 EKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQ 377
EKCESLIIQCL DAKVEV ++NDVI+VGGCSYIP+
Sbjct: 13 EKCESLIIQCLQDAKVEVEEVNDVIIVGGCSYIPR 117
Score = 22.3 bits (46), Expect(2) = 6e-12
Identities = 10/18 (55%), Positives = 13/18 (71%)
Frame = +2
Query: 377 QGWRILLPTYAKLQKFIK 394
+G +ILL TY K + FIK
Sbjct: 113 RG*KILLLTYVKARNFIK 166
>TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (22%)
Length = 769
Score = 62.0 bits (149), Expect = 4e-10
Identities = 58/224 (25%), Positives = 98/224 (42%), Gaps = 14/224 (6%)
Frame = +2
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 104 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 277
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + L PF V +P I + + EE+ +
Sbjct: 278 INTVFDAKRLIGRRFSDASVQSDMKLWPFKV-IPGPADKPMIVVNYKGDEKQFSAEEISS 454
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVP-VSFSRFQLTRL------------ERACAMAGLH 194
M L++++ + E +L I+N + F+ F +L +R C +
Sbjct: 455 MVLIKMKEIAEAYLGSTIKNASCHPSLLYFNDFTSVKLPXGRLVVNXPGFKRGCRIXSXE 634
Query: 195 VLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGY 238
L P +AL+ Q K +G G +++ F G+
Sbjct: 635 --XLTAGPGQLALMGPLXQGKGPLAFLGEGKKRVLQSFEPXVGW 760
>TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat shock cognate
protein 70, partial (54%)
Length = 1266
Score = 54.3 bits (129), Expect = 9e-08
Identities = 24/54 (44%), Positives = 38/54 (69%)
Frame = +1
Query: 329 VDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
V RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 28 VTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQLL 189
>TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight heat shock
protein, partial (54%)
Length = 1272
Score = 53.5 bits (127), Expect = 2e-07
Identities = 22/49 (44%), Positives = 35/49 (70%)
Frame = +1
Query: 329 VDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQ 377
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+
Sbjct: 19 ITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPK 165
>BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chaperone hsp70 -
rice (fragment), partial (18%)
Length = 398
Score = 48.9 bits (115), Expect = 4e-06
Identities = 33/91 (36%), Positives = 49/91 (53%), Gaps = 1/91 (1%)
Frame = -3
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 345 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAK--NQVAMNP 172
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLV 118
+T+F+ KRLIGR +D V + L PF V
Sbjct: 171 INTVFDAKRLIGRRFSDASVQSDIKLWPFKV 79
>AW757289
Length = 402
Score = 42.0 bits (97), Expect = 5e-04
Identities = 18/52 (34%), Positives = 34/52 (64%)
Frame = +3
Query: 331 RAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
RA FEE+N ++F K + + + DA ++ I++++LVGG + IP+ ++L
Sbjct: 12 RARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLL 167
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,643,597
Number of Sequences: 63676
Number of extensions: 173441
Number of successful extensions: 852
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 825
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 827
length of query: 394
length of database: 12,639,632
effective HSP length: 99
effective length of query: 295
effective length of database: 6,335,708
effective search space: 1869033860
effective search space used: 1869033860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC145222.11


