

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145218.2 + phase: 1 /pseudo/partial
(365 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
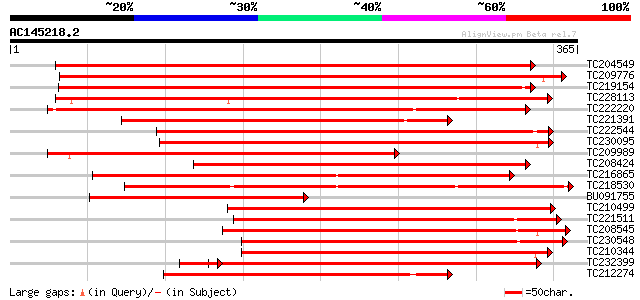
Score E
Sequences producing significant alignments: (bits) Value
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 381 e-106
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 378 e-105
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 370 e-103
TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 347 5e-96
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 330 8e-91
TC221391 similar to UP|O81906 (O81906) Serine/threonine kinase-l... 303 6e-83
TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precu... 289 2e-78
TC230095 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like prot... 275 2e-74
TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 271 3e-73
TC208424 weakly similar to UP|O65466 (O65466) Serine/threonine k... 261 5e-70
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 258 3e-69
TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partia... 257 7e-69
BU091755 253 7e-68
TC210499 weakly similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine k... 240 6e-64
TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, ... 240 8e-64
TC208545 weakly similar to UP|Q9S972 (Q9S972) ARK2 product/recep... 238 3e-63
TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-l... 237 5e-63
TC210344 similar to UP|Q9AVE0 (Q9AVE0) SRKb, partial (21%) 236 2e-62
TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial... 235 2e-62
TC212274 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 227 7e-60
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 381 bits (978), Expect = e-106
Identities = 180/309 (58%), Positives = 237/309 (76%)
Frame = +3
Query: 30 DSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGL 89
D +++ + F +I+AATN FS NKLG+GG+G VYKG GQ +A+KRLS S QG
Sbjct: 765 DIPTVDSLQFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRLSKSSGQGG 944
Query: 90 QEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKL 149
+EFKNE+V++AKLQHRNLVRL G+C++G+EKIL+YEY+ NKSLD +FD + L W
Sbjct: 945 EEFKNEVVVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQRELDWGR 1124
Query: 150 RFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGAS 209
R+ II GIARG+ YLH+DSRLR+IHRDLK SNILLD +M PKISDFG+A+IFG +T +
Sbjct: 1125 RYKIIGGIARGIQYLHEDSRLRIIHRDLKASNILLDGDMNPKISDFGMARIFGVDQTQGN 1304
Query: 210 TQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAW 269
T R++GTYGYM+PEYA+ G FS+KSDV+SFGV+L+EILSGKKN+ F+++ LL YAW
Sbjct: 1305 TSRIVGTYGYMAPEYAMHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTDGAEDLLSYAW 1484
Query: 270 RLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATI 329
+LW + L+LMD L E+ N+NE ++ IGLLCVQ++P +RPTM+ I+ MLD T T+
Sbjct: 1485 QLWKDGTPLELMDPILRESYNQNEVIRSIHIGLLCVQEDPADRPTMATIVLMLDSNTVTL 1664
Query: 330 PIPSQPTFF 338
P P+QP FF
Sbjct: 1665 PTPTQPAFF 1691
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 378 bits (971), Expect = e-105
Identities = 177/338 (52%), Positives = 250/338 (73%), Gaps = 12/338 (3%)
Frame = +2
Query: 33 SIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEF 92
++E + F +I+AAT FS++NKLG+GG+G VYKG P GQE+A+KRLS +S QG +EF
Sbjct: 26 AVESLRFDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEF 205
Query: 93 KNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFD 152
KNE+ ++AKLQHRNLVRL G+C++G+EKIL+YE+++NKSLD +FD + L W R+
Sbjct: 206 KNEVEIVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWTRRYK 385
Query: 153 IIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQR 212
I+ GIARG+ YLH+DSRL++IHRDLK SN+LLD +M PKISDFG+A+IFG +T A+T R
Sbjct: 386 IVEGIARGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNR 565
Query: 213 VMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLW 272
++GTYGYMSPEYA+ G +S KSDV+SFGV++LEI+SGK+N+ F+ + LL YAW+LW
Sbjct: 566 IVGTYGYMSPEYAMHGEYSAKSDVYSFGVLILEIISGKRNSSFYETDVAEDLLSYAWKLW 745
Query: 273 TENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIP 332
+ L+LMD +L E+ NE ++C IGLLCVQ++P +RPTM++++ MLD + T+ +P
Sbjct: 746 KDEAPLELMDQSLRESYTRNEVIRCIHIGLLCVQEDPIDRPTMASVVLMLDSYSVTLQVP 925
Query: 333 SQPTFFTTKH------------QSCSSSSSKLEISMQI 358
+QP F+ QS ++S+SK M +
Sbjct: 926 NQPAFYINSRTEPNMPKGLKIDQSTTNSTSKSVNDMSV 1039
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 370 bits (951), Expect = e-103
Identities = 177/307 (57%), Positives = 234/307 (75%)
Frame = +1
Query: 32 ESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQE 91
+ ++VP + +I AAT+NF +NK+G+GG+GPVYKG+ GGQEIA+KRLSS+S QG+ E
Sbjct: 292 QDVDVPLFDMLTITAATDNFLLNNKIGEGGFGPVYKGKLVGGQEIAVKRLSSLSGQGITE 471
Query: 92 FKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRF 151
F E+ LIAKLQHRNLV+L G CIKG EK+L+YEY+ N SL++FIFD+ ++ LL W RF
Sbjct: 472 FITEVKLIAKLQHRNLVKLLGCCIKGQEKLLVYEYVVNGSLNSFIFDQIKSKLLDWPRRF 651
Query: 152 DIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQ 211
+II+GIARG+LYLHQDSRLR+IHRDLK SN+LLD+++ PKISDFG+A+ FGG +T +T
Sbjct: 652 NIILGIARGLLYLHQDSRLRIIHRDLKASNVLLDEKLNPKISDFGMARAFGGDQTEGNTN 831
Query: 212 RVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRL 271
RV+GTYGYM+PEYA DG FSIKSDVFSFG++LLEI+ G KN Q +L+GYAW L
Sbjct: 832 RVVGTYGYMAPEYAFDGNFSIKSDVFSFGILLLEIVCGIKNKSLCHENQTLNLVGYAWAL 1011
Query: 272 WTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPI 331
W E L L+DS + ++C E ++C + LLCVQ P +RPTM++++ ML E +
Sbjct: 1012WKEQNALQLIDSGIKDSCVIPEVLRCIHVSLLCVQQYPEDRPTMTSVIQMLGSEMDMVE- 1188
Query: 332 PSQPTFF 338
P +P FF
Sbjct: 1189PKEPGFF 1209
>TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (53%)
Length = 1492
Score = 347 bits (890), Expect = 5e-96
Identities = 173/359 (48%), Positives = 242/359 (67%), Gaps = 39/359 (10%)
Frame = +1
Query: 30 DSESIEVPY---YTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVST 86
D + IE+ + F +I+ AT +FSDSNKLGQGG+G VY+GR GQ IA+KRLS S+
Sbjct: 82 DDDEIEIAQSLQFNFDTIRVATEDFSDSNKLGQGGFGAVYRGRLSDGQMIAVKRLSRESS 261
Query: 87 QGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDR------- 139
QG EFKNE++L+AKLQHRNLVRL G+C++G E++L+YEY+ NKSLD FIF R
Sbjct: 262 QGDTEFKNEVLLVAKLQHRNLVRLLGFCLEGKERLLIYEYVPNKSLDYFIFGRS**VHN* 441
Query: 140 -----------------------------TRTVLLGWKLRFDIIVGIARGMLYLHQDSRL 170
T+ L W++R+ II G+ARG+LYLH+DS L
Sbjct: 442 TVALYFTSGSGQRLNIHIPLKLMAVYADPTKKAQLNWEMRYKIITGVARGLLYLHEDSHL 621
Query: 171 RVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFF 230
R+IHRDLK SNILL++EM PKI+DFG+A++ +T A+T R++GTYGYM+PEYA+ G F
Sbjct: 622 RIIHRDLKASNILLNEEMNPKIADFGMARLVLMDQTQANTNRIVGTYGYMAPEYAMHGQF 801
Query: 231 SIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDLMDSALSETCN 290
S+KSDVFSFGV++LEI+SG KN+G + + LL +AWR W E + ++D +L+ +
Sbjct: 802 SMKSDVFSFGVLVLEIISGHKNSGIRHGENVEDLLSFAWRNWREGTAVKIVDPSLNNN-S 978
Query: 291 ENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPTFFTTKHQSCSSSS 349
NE ++C IGLLCVQ+ +RPTM+ I+ ML+ + ++PIPS+P F+ + S++
Sbjct: 979 RNEMLRCIHIGLLCVQENLADRPTMTTIMLMLNSYSLSLPIPSKPAFYVSSRTGSISAT 1155
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 330 bits (845), Expect = 8e-91
Identities = 164/312 (52%), Positives = 223/312 (70%), Gaps = 1/312 (0%)
Frame = +3
Query: 25 NIGENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSV 84
N GE +S ++E + +I+AATN FS ++G+GG+G VYKG P G+EIA+K+LS
Sbjct: 150 NFGE-ESATLESLQFGLVTIEAATNKFSYEKRIGEGGFGVVYKGVLPDGREIAVKKLSKS 326
Query: 85 STQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVL 144
S QG EFKNEI+LIAKLQHRNLV L G+C++ EK+L+YE++SNKSLD F+FD R+
Sbjct: 327 SGQGANEFKNEILLIAKLQHRNLVTLLGFCLEEHEKMLIYEFVSNKSLDYFLFDSHRSKQ 506
Query: 145 LGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGK 204
L W R+ II GIA+G+ YLH+ SRL+VIHRDLK SN+LLD M PKISDFG+A+I
Sbjct: 507 LNWSERYKIIEGIAQGISYLHEHSRLKVIHRDLKPSNVLLDSNMNPKISDFGMARIVAID 686
Query: 205 ETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSL 264
+ T R++GTYGYMSPEYA+ G FS KSDVFSFGV++LEI+S K+NT S L
Sbjct: 687 QLQGKTNRIVGTYGYMSPEYAMHGQFSEKSDVFSFGVIVLEIISAKRNTRSVFSDH-DDL 863
Query: 265 LGYAWRLWTENKLLDLMDSAL-SETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLD 323
L Y W W + L++ D ++ +E C+ +E VKC QIGLLCVQ++P +RP ++ +++ L+
Sbjct: 864 LSYTWEQWMDEAPLNIFDQSIEAEFCDHSEVVKCIQIGLLCVQEKPDDRPKITQVISYLN 1043
Query: 324 GETATIPIPSQP 335
+P+P +P
Sbjct: 1044SSITELPLPKKP 1079
>TC221391 similar to UP|O81906 (O81906) Serine/threonine kinase-like protein
, partial (25%)
Length = 642
Score = 303 bits (777), Expect = 6e-83
Identities = 146/213 (68%), Positives = 181/213 (84%)
Frame = +2
Query: 73 GQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSL 132
G+E+A+KRLS S+QGL+EFKNE+VLIAKLQHRNLVRL G CI+G+EKIL+YEY+ NKSL
Sbjct: 2 GEEVAVKRLSRKSSQGLEEFKNEMVLIAKLQHRNLVRLLGCCIQGEEKILVYEYLPNKSL 181
Query: 133 DTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKI 192
D F+FD + L W RF+II GIARG+LYLH+DSRLR+IHRDLK SNILLD+ M PKI
Sbjct: 182 DCFLFDPVKQTQLDWAKRFEIIEGIARGLLYLHRDSRLRIIHRDLKASNILLDESMNPKI 361
Query: 193 SDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKN 252
SDFGLA+IFGG + A+T RV+GTYGYMSPEYA++G FSIKSDV+SFGV+LLEI+SG+KN
Sbjct: 362 SDFGLARIFGGNQNEANTNRVVGTYGYMSPEYAMEGLFSIKSDVYSFGVLLLEIMSGRKN 541
Query: 253 TGFFRSQQISSLLGYAWRLWTENKLLDLMDSAL 285
T FR SSL+GYAW LW+E ++++L+D +L
Sbjct: 542 TS-FRDTDDSSLIGYAWHLWSEQRVMELVDPSL 637
>TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precursor ,
partial (27%)
Length = 861
Score = 289 bits (739), Expect = 2e-78
Identities = 143/257 (55%), Positives = 189/257 (72%), Gaps = 1/257 (0%)
Frame = +3
Query: 95 EIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDII 154
E+V+I+KLQHRNLVRL G CI+ DE++L+YE+M NKSLD+F+FD + +L WK RF+II
Sbjct: 3 EVVVISKLQHRNLVRLLGCCIERDEQMLVYEFMPNKSLDSFLFDPLQRKILDWKKRFNII 182
Query: 155 VGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIF-GGKETGASTQRV 213
GIARG+LYLH+DSRLR+IHRDLK SNILLDDEM PKISDFGLA+I G + A+T+RV
Sbjct: 183 EGIARGILYLHRDSRLRIIHRDLKASNILLDDEMHPKISDFGLARIVRSGDDDEANTKRV 362
Query: 214 MGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWT 273
+GTYGYM PEYA++G FS KSDV+ V+LLEI+SG++NT F+ ++Q SL YAW+LW
Sbjct: 363 VGTYGYMPPEYAMEGIFSEKSDVYXXXVLLLEIVSGRRNTSFYNNEQSLSLXXYAWKLWN 542
Query: 274 ENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPS 333
E + ++D + + E ++C IGLLCVQ+ RPT+S ++ ML E +P P
Sbjct: 543 EGNIKSIIDLEIQDPMFEKSILRCIHIGLLCVQELTKERPTISTVVXMLISEITHLPPPR 722
Query: 334 QPTFFTTKHQSCSSSSS 350
Q F + Q+C SS S
Sbjct: 723 QXXF--VQKQNCQSSES 767
>TC230095 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase
homolog RK20-1, partial (30%)
Length = 819
Score = 275 bits (703), Expect = 2e-74
Identities = 139/258 (53%), Positives = 180/258 (68%), Gaps = 4/258 (1%)
Frame = +3
Query: 97 VLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVG 156
+LIAKLQH+NLVRL G+C + EKIL+YEY+ NKSLD F+FD + L W RF I+ G
Sbjct: 6 LLIAKLQHKNLVRLVGFCQEDREKILIYEYVPNKSLDHFLFDSQKHRQLTWSERFKIVKG 185
Query: 157 IARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGT 216
IARG+LYLH+DSRL++IHRD+K SN+LLD + PKISDFG+A++ + T RV+GT
Sbjct: 186 IARGILYLHEDSRLKIIHRDIKPSNVLLDYGINPKISDFGMARMVATDQIQGCTNRVVGT 365
Query: 217 YGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENK 276
YGYMSPEYA+ G FS KSDVFSFGV++L+I+SGKKN+ F S ++ LL YAW W +
Sbjct: 366 YGYMSPEYAMHGQFSEKSDVFSFGVMVLDIISGKKNSCSFESCRVDDLLSYAWNNWRDES 545
Query: 277 LLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPT 336
L+DS L E+ NE KC QIGLLCVQ+ P +RPTM I++ L + +P P +P
Sbjct: 546 PYQLLDSTLLESYVPNEVEKCMQIGLLCVQENPDDRPTMGTIVSYLSNPSFEMPFPLEPA 725
Query: 337 FF----TTKHQSCSSSSS 350
FF +H + SSS
Sbjct: 726 FFMHGRMRRHSAEHESSS 779
>TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (32%)
Length = 738
Score = 271 bits (693), Expect = 3e-73
Identities = 140/237 (59%), Positives = 176/237 (74%), Gaps = 10/237 (4%)
Frame = +3
Query: 25 NIGENDSESIEVP----------YYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQ 74
N GE D + E+ + F +I+ ATNNFSD+NKLGQGG+G VYKG GQ
Sbjct: 27 NEGEGDDDEGELANDIKTDDQLLQFEFATIKFATNNFSDANKLGQGGFGIVYKGTLSDGQ 206
Query: 75 EIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDT 134
EIAIKRLS S QG EFKNEI+L +LQHRNLVRL G+C E++L+YE++ NKSLD
Sbjct: 207 EIAIKRLSINSNQGETEFKNEILLTGRLQHRNLVRLLGFCFARRERLLIYEFVPNKSLDY 386
Query: 135 FIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISD 194
FIFD + V L ++R+ II GIARG+LYLH+DSRL V+HRDLKTSNILLD E+ PKISD
Sbjct: 387 FIFDPNKRVNLN*EIRYKIIRGIARGLLYLHEDSRLNVVHRDLKTSNILLDGELNPKISD 566
Query: 195 FGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKK 251
FG+A++F +T AST ++GT+GYM+PEY G FSIKSDVFSFGV++LEI+ G++
Sbjct: 567 FGMARLFEINQTEASTTTIVGTFGYMAPEYIKHGQFSIKSDVFSFGVMILEIVCGQR 737
>TC208424 weakly similar to UP|O65466 (O65466) Serine/threonine kinase-like
protein , partial (34%)
Length = 913
Score = 261 bits (666), Expect = 5e-70
Identities = 124/217 (57%), Positives = 163/217 (74%)
Frame = +2
Query: 119 EKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLK 178
EKIL+YEY++NKSLD F+FD + L W R+ IIVGIARG+ YLH+DS+LR+IHRDLK
Sbjct: 2 EKILIYEYITNKSLDHFLFDPVKQKELDWSRRYKIIVGIARGIQYLHEDSQLRIIHRDLK 181
Query: 179 TSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFS 238
SN+LLD+ M PKISDFG+AKIF +T +T R++GTYGYMSPEYA+ G FS+KSDVFS
Sbjct: 182 ASNVLLDENMNPKISDFGMAKIFQADQTQVNTGRIVGTYGYMSPEYAMRGQFSVKSDVFS 361
Query: 239 FGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCA 298
FGV++LEI+SGKKNT F++S LL +AW+ WT L+L+D L + + NE +C
Sbjct: 362 FGVLVLEIVSGKKNTDFYQSNHADDLLSHAWKNWTLQTPLELLDPTLRGSYSRNEVNRCI 541
Query: 299 QIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQP 335
IGLLCVQ+ P +RP+M+ I ML+ + T+ +P QP
Sbjct: 542 HIGLLCVQENPSDRPSMATIALMLNSYSVTMSMPQQP 652
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 258 bits (659), Expect = 3e-69
Identities = 129/273 (47%), Positives = 191/273 (69%), Gaps = 1/273 (0%)
Frame = +1
Query: 54 SNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGY 113
+NK+G+GG+GPVYKG F G IA+K+LSS S QG +EF NEI +I+ LQH +LV+L G
Sbjct: 4 ANKIGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQGNREFLNEIGMISALQHPHLVKLYGC 183
Query: 114 CIKGDEKILLYEYMSNKSLDTFIFD-RTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRV 172
C++GD+ +L+YEYM N SL +F + L W R+ I VGIARG+ YLH++SRL++
Sbjct: 184 CVEGDQLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLKI 363
Query: 173 IHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSI 232
+HRD+K +N+LLD ++ PKISDFGLAK+ T ST R+ GT+GYM+PEYA+ G+ +
Sbjct: 364 VHRDIKATNVLLDQDLNPKISDFGLAKLDEEDNTHIST-RIAGTFGYMAPEYAMHGYLTD 540
Query: 233 KSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDLMDSALSETCNEN 292
K+DV+SFG+V LEI++G+ NT + ++ S+L +A L + ++DL+D L N+
Sbjct: 541 KADVYSFGIVALEIINGRSNTIHRQKEESFSVLEWAHLLREKGDIMDLVDRRLGLEFNKE 720
Query: 293 EFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGE 325
E + ++ LLC RPTMS++++ML+G+
Sbjct: 721 EALVMIKVALLCTNVTAALRPTMSSVVSMLEGK 819
>TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partial (25%)
Length = 1190
Score = 257 bits (656), Expect = 7e-69
Identities = 139/290 (47%), Positives = 196/290 (66%), Gaps = 1/290 (0%)
Frame = +3
Query: 75 EIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDT 134
EIA+K+LS S QG +F EI I+ +QHRNLV+L G CI+G +++L+YEY+ NKSLD
Sbjct: 3 EIAVKQLSVGSHQGKSQFITEIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQ 182
Query: 135 FIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISD 194
+F + T L W R+DI +G+ARG+ YLH++SRLR++HRD+K SNILLD E+IPKISD
Sbjct: 183 ALFGKCLT--LNWSTRYDICLGVARGLTYLHEESRLRIVHRDVKASNILLDYELIPKISD 356
Query: 195 FGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTG 254
FGLAK++ K+T ST V GT GY++PEYA+ G + K+DVFSFGVV LE++SG+ N+
Sbjct: 357 FGLAKLYDDKKTHIST-GVAGTIGYLAPEYAMRGHLTEKADVFSFGVVALELVSGRPNSD 533
Query: 255 FFRSQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPT 314
+ LL +AW+L +N ++DL+D LSE NE E + I LLC Q P RP+
Sbjct: 534 SSLEGEKVYLLEWAWQLHEKNCIIDLVDDRLSE-FNEEEVKRVVGIALLCTQTSPTLRPS 710
Query: 315 MSNILTMLDGETATIPIPSQPTFFTT-KHQSCSSSSSKLEISMQIDSSYQ 363
MS ++ ML G+ + S+P + + K + SS + +EI D++YQ
Sbjct: 711 MSRVVAMLSGDIEVSTVTSKPGYLSDWKFEDVSSFMTGIEIKGS-DTNYQ 857
>BU091755
Length = 424
Score = 253 bits (647), Expect = 7e-68
Identities = 123/141 (87%), Positives = 134/141 (94%)
Frame = +1
Query: 52 SDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLR 111
+DS KLG+GGYGPVYKG FPGGQ+IA+KRLSSVSTQGL+EFKNE++LIAKLQHRNLVRLR
Sbjct: 1 TDSYKLGRGGYGPVYKGTFPGGQDIAVKRLSSVSTQGLEEFKNEVILIAKLQHRNLVRLR 180
Query: 112 GYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLR 171
GYCIKGDEKILLYEYM NKSLD+FIFDRTRT LL W +RF+IIVGIARGMLYLHQDSRLR
Sbjct: 181 GYCIKGDEKILLYEYMPNKSLDSFIFDRTRTSLLDWPIRFEIIVGIARGMLYLHQDSRLR 360
Query: 172 VIHRDLKTSNILLDDEMIPKI 192
VIHRDLKTSNILLD+EM PKI
Sbjct: 361 VIHRDLKTSNILLDEEMNPKI 423
>TC210499 weakly similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine kinase-like
protein , partial (27%)
Length = 717
Score = 240 bits (613), Expect = 6e-64
Identities = 110/211 (52%), Positives = 161/211 (76%)
Frame = +3
Query: 141 RTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKI 200
R++LL WK RF+II GI++G+LYLH+ SRL++IHRDLK SNILLD+ M PKISDFGLA++
Sbjct: 3 RSMLLDWKKRFNIIEGISQGILYLHKYSRLKIIHRDLKASNILLDENMNPKISDFGLARM 182
Query: 201 FGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQ 260
F +E+ +T R++GTYGYMSPEYA++G FS KSDV+SFGV+LLEI+SG+KNT F+
Sbjct: 183 FMQQESTGTTSRIVGTYGYMSPEYAMEGTFSTKSDVYSFGVLLLEIVSGRKNTSFYDVDH 362
Query: 261 ISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILT 320
+ +L+G+AW LW + + L L+D +L+++ + +E +C +GLLCV+ +RPTMSN+++
Sbjct: 363 LLNLIGHAWELWNQGESLQLLDPSLNDSFDPDEVKRCIHVGLLCVEHYANDRPTMSNVIS 542
Query: 321 MLDGETATIPIPSQPTFFTTKHQSCSSSSSK 351
ML E+A + +P +P F+ + +SSK
Sbjct: 543 MLTNESAPVTLPRRPAFYVERKNFDGKTSSK 635
>TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, partial
(21%)
Length = 765
Score = 240 bits (612), Expect = 8e-64
Identities = 112/211 (53%), Positives = 156/211 (73%)
Frame = +3
Query: 145 LGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGK 204
L W RF+II GIA+G+LYLHQDSRLR+IHRDLK SN+LLD E+ PKISDFG+A+IFG
Sbjct: 3 LDWSKRFNIICGIAKGLLYLHQDSRLRIIHRDLKASNVLLDSELNPKISDFGMARIFGVD 182
Query: 205 ETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSL 264
+ +T+R++GTYGYM+PEYA DG FS+KSDVFSFGV+LLEI+SGK++ G++ +L
Sbjct: 183 QQEGNTKRIVGTYGYMAPEYATDGLFSVKSDVFSFGVLLLEIISGKRSRGYYNQNHSQNL 362
Query: 265 LGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDG 324
+G+AW+LW E + L+L+D ++ ++ + ++ + C + LLCVQ P +RP MS++L ML
Sbjct: 363 IGHAWKLWKEGRPLELIDKSIEDSSSLSQMLHCIHVSLLCVQQNPEDRPGMSSVLLMLVS 542
Query: 325 ETATIPIPSQPTFFTTKHQSCSSSSSKLEIS 355
E +P P QP FF SS+SK ++S
Sbjct: 543 E-LELPEPKQPGFFGKYSGEADSSTSKQQLS 632
>TC208545 weakly similar to UP|Q9S972 (Q9S972) ARK2 product/receptor-like
serine/threonine protein kinase ARK2, partial (20%)
Length = 1253
Score = 238 bits (607), Expect = 3e-63
Identities = 117/233 (50%), Positives = 166/233 (71%), Gaps = 9/233 (3%)
Frame = +2
Query: 138 DRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGL 197
D TR+ LL WK RF+II GI++G+LYLH+ SRL+VIHRDLK SNILLD+ M PKISDFGL
Sbjct: 326 DGTRSKLLDWKKRFNIIEGISQGLLYLHKYSRLKVIHRDLKASNILLDENMNPKISDFGL 505
Query: 198 AKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFR 257
A++F +E+ +T R++GTYGYMSPEYA++G FS+KSDV+SFGV+LLEI+SG++NT F+
Sbjct: 506 ARMFTRQESTTNTSRIVGTYGYMSPEYAMEGVFSVKSDVYSFGVLLLEIVSGRRNTSFYD 685
Query: 258 SQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSN 317
+ +L+G+AW LW E L L+D +L+E+ + +E +C IGLLCV+ NRP MS
Sbjct: 686 GDRFLNLIGHAWELWNEGACLKLIDPSLTESPDLDEVQRCIHIGLLCVEQNANNRPLMSQ 865
Query: 318 ILTMLDGETATIPIPSQPTFF---------TTKHQSCSSSSSKLEISMQIDSS 361
I++ML + I +P +P F+ + + C+ S+ + S +I+SS
Sbjct: 866 IISMLSNKN-PITLPQRPAFYFGSETFDGIISSTEFCTDSTKAITTSREIESS 1021
>TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-like kinase
RLK14, partial (20%)
Length = 692
Score = 237 bits (605), Expect = 5e-63
Identities = 116/210 (55%), Positives = 153/210 (72%)
Frame = +3
Query: 150 RFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGAS 209
RF II GIARG+LYLHQDSRLR+IHRDLK SN+LLD+EM PKISDFGLA++ GG +
Sbjct: 3 RFGIINGIARGLLYLHQDSRLRIIHRDLKASNVLLDNEMNPKISDFGLARMCGGDQIEGE 182
Query: 210 TQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAW 269
T RV+GTYGYM+PEYA DG FSIKSDVFSFGV+LLEI+SGKKN+ F ++L+G+AW
Sbjct: 183 TSRVVGTYGYMAPEYAFDGIFSIKSDVFSFGVLLLEIVSGKKNSRLFYPNDYNNLIGHAW 362
Query: 270 RLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATI 329
LW E + +DS+L ++C E ++C IGLLCVQ P +RP M++++ +L E A +
Sbjct: 363 MLWKEGNPMQFIDSSLEDSCILYEALRCIHIGLLCVQHHPNDRPNMASVVVLLSNENA-L 539
Query: 330 PIPSQPTFFTTKHQSCSSSSSKLEISMQID 359
P+P P++ + + SSSK S+ I+
Sbjct: 540 PLPKDPSYLSKDISTERESSSKNFTSVSIN 629
>TC210344 similar to UP|Q9AVE0 (Q9AVE0) SRKb, partial (21%)
Length = 870
Score = 236 bits (601), Expect = 2e-62
Identities = 113/204 (55%), Positives = 150/204 (73%), Gaps = 4/204 (1%)
Frame = +2
Query: 150 RFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGAS 209
R DII GIARG+LYLH+DSRL++IHRDLK SN+LLD +M PKISDFG+A+IF G E A+
Sbjct: 8 RLDIINGIARGILYLHEDSRLKIIHRDLKASNVLLDYDMNPKISDFGMARIFAGSEGEAN 187
Query: 210 TQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAW 269
T ++GTYGYM+PEYA++G +SIKSDVF FGV+LLEI++GK+N GF+ S + SLL YAW
Sbjct: 188 TATIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGKRNAGFYHSNKTPSLLSYAW 367
Query: 270 RLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATI 329
LW E K L+L+D ++C +EF++ IGLLCVQ++ +RPTMS+++ ML E+AT+
Sbjct: 368 HLWNEGKGLELIDPMSVDSCPGDEFLRYMHIGLLCVQEDAYDRPTMSSVVLMLKNESATL 547
Query: 330 PIPSQPTF----FTTKHQSCSSSS 349
P +P F F C S
Sbjct: 548 GQPERPPFSLGRFNANEPDCQDCS 619
>TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial (23%)
Length = 1167
Score = 235 bits (600), Expect = 2e-62
Identities = 108/214 (50%), Positives = 156/214 (72%)
Frame = +2
Query: 129 NKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEM 188
N +L+ + D +++ LL W+ R II GI RG+LYLH+DSRL++IHRDLK SN+LL + +
Sbjct: 299 NATLNVYFLDPSKSKLLDWRKRCGIIEGIGRGLLYLHRDSRLKIIHRDLKASNVLLYEAL 478
Query: 189 IPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILS 248
PKISDFG+A+IFGG E A+T RV+GTYGYMSPEYA+ G FS KSDVFSFGV+++EI+S
Sbjct: 479 NPKISDFGMARIFGGTEDQANTNRVVGTYGYMSPEYAMQGLFSEKSDVFSFGVLVIEIVS 658
Query: 249 GKKNTGFFRSQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDE 308
G++N+ F+ SLLG+AW W E +L ++D + + + + ++C IGLLCVQ+
Sbjct: 659 GRRNSRFYDDDNALSLLGFAWIQWREGNILSVIDPEIYDVTHHKDILRCIHIGLLCVQER 838
Query: 309 PGNRPTMSNILTMLDGETATIPIPSQPTFFTTKH 342
+RPTM+ +++ML+ E A +P P QP F +++
Sbjct: 839 AVDRPTMAAVISMLNSEVAFLPPPDQPAFVQSQN 940
Score = 45.4 bits (106), Expect = 4e-05
Identities = 20/28 (71%), Positives = 23/28 (81%)
Frame = +3
Query: 110 LRGYCIKGDEKILLYEYMSNKSLDTFIF 137
L G C +GDEK+L+YEYM NKSLD FIF
Sbjct: 3 LFGCCAEGDEKMLIYEYMLNKSLDVFIF 86
>TC212274 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (20%)
Length = 553
Score = 227 bits (578), Expect = 7e-60
Identities = 109/186 (58%), Positives = 145/186 (77%)
Frame = +2
Query: 100 AKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIAR 159
AKLQHRNLVRL GYCI+ +E++L+YE+M++KSLD+FIFD +++LL R II G+AR
Sbjct: 2 AKLQHRNLVRLLGYCIQSEERLLVYEFMASKSLDSFIFDENKSMLLDRPTRSLIINGVAR 181
Query: 160 GMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGY 219
G+LYLHQDSR ++HRDLK N+LLD EM PKISD GLA+ FGG E A+T+ V+GTYGY
Sbjct: 182 GLLYLHQDSRHTIVHRDLKAGNVLLDSEMNPKISDSGLARSFGGNEIEATTKHVVGTYGY 361
Query: 220 MSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLD 279
+ PEY +DG +S KSDVFSFGV++LEI+SGK+N GF +LL + WRL+TE K +
Sbjct: 362 LPPEYIIDGAYSTKSDVFSFGVLILEIVSGKRNKGFCHQ---DNLLAHVWRLFTEGKCSE 532
Query: 280 LMDSAL 285
++D+ +
Sbjct: 533 IVDATI 550
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,815,525
Number of Sequences: 63676
Number of extensions: 196680
Number of successful extensions: 2794
Number of sequences better than 10.0: 1013
Number of HSP's better than 10.0 without gapping: 2070
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2100
length of query: 365
length of database: 12,639,632
effective HSP length: 99
effective length of query: 266
effective length of database: 6,335,708
effective search space: 1685298328
effective search space used: 1685298328
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC145218.2


