

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144766.5 + phase: 0
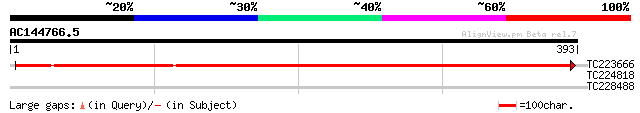
(393 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC223666 UP|O81532 (O81532) Mariner transposase, complete 434 e-122
TC224818 UP|Q6S4R7 (Q6S4R7) ADP-ribosylation factor, complete 28 5.5
TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%) 28 7.2
>TC223666 UP|O81532 (O81532) Mariner transposase, complete
Length = 1278
Score = 434 bits (1116), Expect = e-122
Identities = 215/390 (55%), Positives = 294/390 (75%), Gaps = 2/390 (0%)
Frame = +1
Query: 5 VASQFSLHIRYVQRIWKLAENKGVHADVSSKRAGNCGRKRV-VDLDRVRDIPLKKRTTIR 63
VAS FS+ + + RIWK A+ H DVS K+ N GRKRV +DL ++R+IPL +RTT+R
Sbjct: 109 VASSFSVCRKTIDRIWKRAKESETH-DVSHKKTKNSGRKRVEIDLSQLREIPLSQRTTVR 285
Query: 64 DLSEALDVSRGTLSRCIRSKEIRKNSNAIKPALTE-GNKARLQFCISMLDSASIDHDPIF 122
L+ A+ + + R I+S I+++S+AIKP LTE G + RL+FC+SML+ I HDP+F
Sbjct: 286 TLAVAMKTNTSAMYRLIQSGAIKRHSSAIKPQLTEEGKRLRLEFCLSMLEG--IPHDPMF 459
Query: 123 KGMYNTVHIDEKWFYLKEKSKKFYLVHDEEDPIFSCQSKNYIPKIMFLVAIARPRFDAQG 182
+ MYN +HIDEKWFY+ +KS+++YL+ DE+ P SC+SKN++PK+MFL A+ARPRFD++
Sbjct: 460 QSMYNIIHIDEKWFYMTKKSERYYLLPDEDKPHRSCKSKNFVPKVMFLTAVARPRFDSEK 639
Query: 183 KEIFSGKIGAFPLITREPARRSSVNRPAGTLETKPIQLITRNVIKKCVLEQVVPSMMELW 242
FSGKIG FP +T+EPA+R+SVNR AGT+ETK I I R++I+ +E+V+P+ E+W
Sbjct: 640 NVTFSGKIGIFPFVTQEPAKRTSVNRVAGTMETKAITSINRDLIRSVFIEKVLPATKEVW 819
Query: 243 PIEERGQPIFIQQDNAKTHIDSEDQDFRCVASQNGFDIRLVRQPPNSPDLNVLDLGFFRS 302
P +E G IFIQQDNA+THI+ +D +F A+Q+GFDIRL+ QPPNSPD NVLDLGFF +
Sbjct: 820 PRDELGSTIFIQQDNARTHINPDDPEFVQAATQDGFDIRLMCQPPNSPDFNVLDLGFFSA 999
Query: 303 IQALQHKKSPNSVDDLVDAVQKSFEEYDTILSNRIFLSLQSCMIEIMKVKGTHNYKIPHM 362
IQ+L +K++P ++D+LV+AV KSFE Y + SN IFLSLQ CMIE MK KG++ Y HM
Sbjct: 1000IQSLHYKEAPKTIDELVNAVVKSFENYCVVKSNFIFLSLQLCMIETMKAKGSNRYTSQHM 1179
Query: 363 KKEQLERRKELPTQRKCDIQLVQEVLSYLD 392
+KE+LE ++LP Q KCD LVQE L YL+
Sbjct: 1180QKEKLETEEQLPIQLKCDPILVQETLDYLN 1269
>TC224818 UP|Q6S4R7 (Q6S4R7) ADP-ribosylation factor, complete
Length = 1067
Score = 28.5 bits (62), Expect = 5.5
Identities = 25/118 (21%), Positives = 47/118 (39%), Gaps = 16/118 (13%)
Frame = -3
Query: 135 WFYLKEKSKKFYLVHDEEDPIFSCQSKNYIPKIMFLVAIARPRFDAQGKEIFSG------ 188
W Y++ K K+ + + C+ NY K + AI + + + + + +
Sbjct: 936 WKYIQSKIKQTRCKQTQHERTRGCKRSNYS*KY*LI*AIIQTQVTQKKRILLASILLLQH 757
Query: 189 ---KIGAFPL-------ITREPARRSSVNRPAGTLETKPIQLITRNVIKKCVLEQVVP 236
KI F ITR+ S+V+ AG + KP+Q + + + C +P
Sbjct: 756 SLEKISIFLQEPPDSHEITRQDQNFSNVSCFAGNVVRKPVQTLIETLSRSCTSALDIP 583
>TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%)
Length = 1303
Score = 28.1 bits (61), Expect = 7.2
Identities = 15/64 (23%), Positives = 30/64 (46%)
Frame = +1
Query: 213 LETKPIQLITRNVIKKCVLEQVVPSMMELWPIEERGQPIFIQQDNAKTHIDSEDQDFRCV 272
L T P + T +++K+C L Q + ++ E WP + + + + Q + ++F
Sbjct: 946 LMTLPSRCGTLDMVKQCQLLQTIKNLFEQWPSIPKSKLLHLLQLTILKSSTFQKENFATT 1125
Query: 273 ASQN 276
S N
Sbjct: 1126 CSLN 1137
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,856,434
Number of Sequences: 63676
Number of extensions: 202394
Number of successful extensions: 861
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 857
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 858
length of query: 393
length of database: 12,639,632
effective HSP length: 99
effective length of query: 294
effective length of database: 6,335,708
effective search space: 1862698152
effective search space used: 1862698152
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144766.5


