

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.7 - phase: 0
(482 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
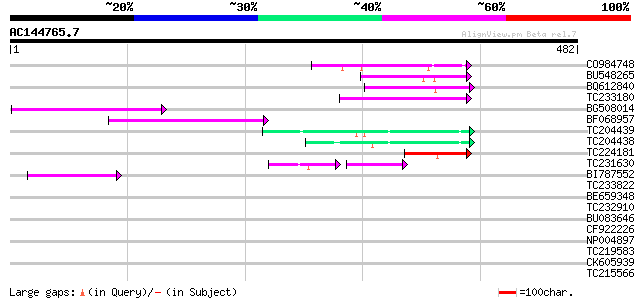
Score E
Sequences producing significant alignments: (bits) Value
CO984748 76 3e-14
BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delal... 73 2e-13
BQ612840 63 3e-10
TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, parti... 62 7e-10
BG508014 GP|15128578|db prolactin receptor {Paralichthys olivace... 59 6e-09
BF068957 weakly similar to PIR|B85188|B85 retrotransposon like p... 50 2e-06
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 50 3e-06
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 49 5e-06
TC224181 48 1e-05
TC231630 weakly similar to UP|Q7PR65 (Q7PR65) ENSANGP00000016899... 36 6e-05
BI787552 45 9e-05
TC233822 40 0.003
BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor p... 39 0.005
TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial ... 30 0.015
BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 37 0.020
CF922226 37 0.026
NP004897 gag-protease polyprotein 34 0.13
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 32 0.48
CK605939 32 0.63
TC215566 similar to UP|Q9FSG4 (Q9FSG4) Wound-induced GSK-3-like ... 32 0.63
>CO984748
Length = 810
Score = 76.3 bits (186), Expect = 3e-14
Identities = 55/168 (32%), Positives = 82/168 (48%), Gaps = 32/168 (19%)
Frame = -2
Query: 257 KAPCQVCGKTNHTAINCFHRFDKNY-------------------SRSNYSADSDKQGSHN 297
K CQ K +H A +C+HRF+ NY RSN A ++K S
Sbjct: 806 KIQCQXXYKFDHDAADCYHRFNPNYVAQQYNQNQSLNQHRNYSYQRSNCQALNNKSQSQQ 627
Query: 298 ------AFIASQNSVED-YDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEI 350
A++A+ N + +WY + GA++HVT + G + +GNG L I
Sbjct: 626 NAQAPQAYLANANPTQQSQNWYINYGATHHVTASPQNLMNEVPTSGNEQVFLGNGQGLPI 447
Query: 351 VATC------SSKLKSLNLDDVLYVPNITKNLLSVSKLAADNNIFVEF 392
+ +S +K L L+++L+VP+ITKNL+SVS A D N+F EF
Sbjct: 446 TGSTVFNSPFASNVK-LTLNNLLHVPHITKNLVSVS*FAKD-NVFFEF 309
>BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delalandii},
partial (5%)
Length = 667
Score = 73.2 bits (178), Expect = 2e-13
Identities = 39/101 (38%), Positives = 58/101 (56%), Gaps = 7/101 (6%)
Frame = -3
Query: 299 FIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEI----VATC 354
F+ S + WY DSGAS+HVT+ + Q + +++GNG L I ++T
Sbjct: 653 FLTSSAATPSQSWYPDSGASHHVTNMSQNIQQVAPFEEPIQIIIGNGQGLNINSSGLSTF 474
Query: 355 SSKLK---SLNLDDVLYVPNITKNLLSVSKLAADNNIFVEF 392
SS + SL L ++L+VP ITKNL+ VS+ DNN++ EF
Sbjct: 473 SSPINPQFSLVLSNLLFVPTITKNLIRVSQFCKDNNVYFEF 351
>BQ612840
Length = 441
Score = 62.8 bits (151), Expect = 3e-10
Identities = 38/101 (37%), Positives = 53/101 (51%), Gaps = 7/101 (6%)
Frame = +2
Query: 302 SQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLKS- 360
SQ + W DSGAS H ++ Q + G + + +GNG L+I ++ SS S
Sbjct: 41 SQPNSNSVIWIPDSGASCHFIGESQNIQQIEPFEGIDRIFIGNGQDLKITSSGSSSFISP 220
Query: 361 ------LNLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKN 395
L+L+ L+VP ITKNL+SVS+ A DN EF N
Sbjct: 221 YNSKVFLSLNRFLHVPYITKNLVSVSQFARDNGFCFEFHSN 343
>TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, partial (11%)
Length = 916
Score = 61.6 bits (148), Expect = 7e-10
Identities = 35/113 (30%), Positives = 63/113 (54%), Gaps = 1/113 (0%)
Frame = +2
Query: 281 YSRSNYSADSDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSL 340
+S+++ S G ++F++ S + DSG ++H+T ++ F T G +
Sbjct: 362 FSKTSSSCSLTMAGKSSSFLSFNASGTENI*IIDSGVTDHMTPHSSYFSSYTFLIGNQHI 541
Query: 341 VVGNGDKLEIVATCSSKLKS-LNLDDVLYVPNITKNLLSVSKLAADNNIFVEF 392
+V NG + I+ + +L+S L+L++VLYVP ++ NLLS+ K+ D N V F
Sbjct: 542 IVANGSHIPIIGCGNIQLQSSLHLNNVLYVPKLSNNLLSIHKIT*DLNCVVTF 700
>BG508014 GP|15128578|db prolactin receptor {Paralichthys olivaceus}, partial
(2%)
Length = 450
Score = 58.5 bits (140), Expect = 6e-09
Identities = 32/132 (24%), Positives = 63/132 (47%)
Frame = +1
Query: 2 ASAANNNKNDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDS 61
+S++ + S+S KL NY LW V V++ L +++ + +
Sbjct: 37 SSSSPTSLKQFSHSISHKLHVTNYLLWLQQVELVIKSRHLHHFLVNPLIPKKYALVKDCD 216
Query: 62 SKSNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQII 121
S + + W+ DQ L W+ +++ +M T ++ C++S QLWD Q + TR+++
Sbjct: 217 SDTTTEVYHGWKEQDQFFLAWLRSTIYGDMLTHVIGCKSSWQLWDRLQLHFQSLTRARVR 396
Query: 122 YLKSEFHSIRKG 133
L++E + G
Sbjct: 397 QLRNELRCLSHG 432
>BF068957 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (3%)
Length = 420
Score = 50.1 bits (118), Expect = 2e-06
Identities = 32/137 (23%), Positives = 69/137 (50%), Gaps = 1/137 (0%)
Frame = -2
Query: 85 NSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRKGEMKMEDYLIKM 144
++++ + +++L + S Q+WD+ HT+++ L + ++ +E+ L K+
Sbjct: 413 STLSKSVLSRVLASDHSYQVWDKIHEHFSLHTKTRAPQLHTAMRAVTLDGKTIEE*L*KI 234
Query: 145 KNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIV-VKLSDHTTLSWVDLQAQLLTFESR 203
K ++L G P+ + + + L GL S+Y +V V S T S +++A L E+R
Sbjct: 233 KGFVNELASVGVPVHHKEYVEALLEGLPSDYASVVSVIESKKRTPSIAEIEALLYRHETR 54
Query: 204 IEQLNNLTNLNLNATAN 220
+ + N + +A+ N
Sbjct: 53 LMRYNKEAQVMNSASIN 3
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 49.7 bits (117), Expect = 3e-06
Identities = 49/188 (26%), Positives = 75/188 (39%), Gaps = 8/188 (4%)
Frame = +1
Query: 216 NATANVANKFDHRDNRFNSNNNWRGSNFRGWRGGRGRGRSSKAPCQVCGKTNHTAINCFH 275
+A +F NR + + S G + + + + K C CGK H C+H
Sbjct: 1384 SAGRTTMTEFVPAKNRTGATMSQHRSRHHGMQ--QKKSKRKKWRCHYCGKYGHIKPFCYH 1557
Query: 276 RFDKNYSRSNYSADSDKQ---GSHNAFI----ASQNSVEDYDWYFDSGASNHVTHQTNKF 328
+ + S K H A S + DWY DSG S H+T +F
Sbjct: 1558 LHGHPHHGTQSSNSRKKMMWVPKHKAVSLVVHTSLRASAKEDWYLDSGCSRHMT-GVKEF 1734
Query: 329 QDLTEHHGKNSLVVGNGDKLEIVATCSSKLKSL-NLDDVLYVPNITKNLLSVSKLAADNN 387
E + + G+G K +I+ L +L+ VL V +T NL+S+S+L D
Sbjct: 1735 LLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQL-CDEG 1911
Query: 388 IFVEFDKN 395
V F K+
Sbjct: 1912 FNVNFTKS 1935
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 48.9 bits (115), Expect = 5e-06
Identities = 41/156 (26%), Positives = 62/156 (39%), Gaps = 12/156 (7%)
Frame = +1
Query: 252 RGRSSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIASQNSVE---- 307
+ + K C CGK H C+H + ++ S G ++ V
Sbjct: 1489 KSKRKKWRCHYCGKYGHIKPFCYHL----HGHPHHGTQSSSSGRKMMWVPKHKIVSLVVH 1656
Query: 308 -------DYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLKS 360
DWY DSG S H+T +F E + + G+G K +I
Sbjct: 1657 TSLRASAKEDWYLDSGCSRHMT-GVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHDG 1833
Query: 361 L-NLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKN 395
L +L+ VL V +T NL+S+S+L D V F K+
Sbjct: 1834 LPSLNKVLLVKGLTANLISISQL-CDEGFNVNFTKS 1938
>TC224181
Length = 597
Score = 47.8 bits (112), Expect = 1e-05
Identities = 28/64 (43%), Positives = 40/64 (61%), Gaps = 7/64 (10%)
Frame = +2
Query: 336 GKNSLVVGNGDKLEIVATCSSKLKS-LN------LDDVLYVPNITKNLLSVSKLAADNNI 388
G + + +GN L I + S KS LN L ++L+VP+ITKNLLSVS+ A DN++
Sbjct: 158 GPDQIFIGNSQGLHIKGSSCSFFKSPLNSKFSFVLKNLLHVPSITKNLLSVSQFARDNSV 337
Query: 389 FVEF 392
+ EF
Sbjct: 338 YFEF 349
>TC231630 weakly similar to UP|Q7PR65 (Q7PR65) ENSANGP00000016899 (Fragment),
partial (13%)
Length = 709
Score = 35.8 bits (81), Expect(2) = 6e-05
Identities = 25/73 (34%), Positives = 33/73 (44%), Gaps = 12/73 (16%)
Frame = -2
Query: 221 VANKFDHRD-NRFNSNNNWRGSNFRGWRGGRGR-----------GRSSKAPCQVCGKTNH 268
V NK D + F + GS+ G RGG GR GR + CQVC K H
Sbjct: 531 VPNKTDQESFSAFRGGSGCGGSSPHG-RGGVGRRGGGSGCGRRGGRFANFQCQVCFKFVH 355
Query: 269 TAINCFHRFDKNY 281
T C++ +D+ Y
Sbjct: 354 TTSVCYYHYDQYY 316
Score = 28.9 bits (63), Expect(2) = 6e-05
Identities = 16/53 (30%), Positives = 25/53 (46%), Gaps = 1/53 (1%)
Frame = -1
Query: 287 SADSDKQGSHNAFIA-SQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKN 338
S ++ Q + A + +Q +W +SGAS HVT ++ Q L G N
Sbjct: 229 STSTNHQSNPGAMLTDAQPQSNSNNWIPNSGASFHVTRESRNIQQLEPFEGPN 71
>BI787552
Length = 421
Score = 44.7 bits (104), Expect = 9e-05
Identities = 22/80 (27%), Positives = 40/80 (49%)
Frame = +2
Query: 16 VSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQAN 75
V LD NY W+ V PV++ KL +++ P F + NP +E W+
Sbjct: 182 VQENLDDLNYLHWRQHVEPVIKLHKLQRFVVNLVVPPRYFTEDDRIADRVNPEYETWEVQ 361
Query: 76 DQRLLGWMLNSMATEMATQL 95
DQ LL W+ ++++ + +++
Sbjct: 362 DQTLLVWLQSTLSKSVLSRV 421
>TC233822
Length = 632
Score = 39.7 bits (91), Expect = 0.003
Identities = 26/81 (32%), Positives = 41/81 (50%), Gaps = 1/81 (1%)
Frame = +2
Query: 314 DSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCS-SKLKSLNLDDVLYVPNI 372
DS AS+H+ ++ F + + + + NG K+ S SLNLD + +VPN
Sbjct: 44 DSTASDHIFGNSSLFTSQSPPKIPHLITLANGTKVTSKGFGKFSIFPSLNLDPIHFVPNC 223
Query: 373 TKNLLSVSKLAADNNIFVEFD 393
NL+S+S+L N + FD
Sbjct: 224 PFNLVSLSQLTKALNFSITFD 286
>BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor protein
DSK-R1 - fruit fly (Drosophila melanogaster), partial
(6%)
Length = 770
Score = 38.9 bits (89), Expect = 0.005
Identities = 36/140 (25%), Positives = 58/140 (40%), Gaps = 27/140 (19%)
Frame = -1
Query: 236 NNWRGSNFRGWRGGRGRGRSSKAPCQVCGKTNHTAINCF--HRF---DKNYSRSNYS--- 287
+N G RG +GGR GR + C C + HT C+ H F N S+S S
Sbjct: 452 SNRGGQGGRGNQGGRA-GRGGRP*CSYCKRVGHTQDTCYSIHGFPGKSVNISKSETSEIK 276
Query: 288 ---AD--------SDKQGSHNAFIASQNSV--------EDYDWYFDSGASNHVTHQTNKF 328
AD + K+ ++ I+ NS W DSGAS+H+ ++ F
Sbjct: 275 FLEADYQEYLQLKATKESQTSSVISGHNSTACISQVGNNQSPWIIDSGASDHIASNSSLF 96
Query: 329 QDLTEHHGKNSLVVGNGDKL 348
L+ + + + +G ++
Sbjct: 95 *FLSPPKIPHFITLADGSRV 36
>TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial (3%)
Length = 690
Score = 29.6 bits (65), Expect(2) = 0.015
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = +1
Query: 300 IASQNSVEDYDWYFDSGASNHVTHQTNKF 328
I N +D W+FDSGA++HV F
Sbjct: 121 ILKSNKDDDVAWWFDSGATSHVCKDRRLF 207
Score = 26.6 bits (57), Expect(2) = 0.015
Identities = 15/20 (75%), Positives = 16/20 (80%)
Frame = +2
Query: 359 KSLNLDDVLYVPNITKNLLS 378
KSL LD VL+VP I KNLLS
Sbjct: 314 KSLYLD-VLFVPGIRKNLLS 370
>BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (19%)
Length = 428
Score = 37.0 bits (84), Expect = 0.020
Identities = 28/90 (31%), Positives = 43/90 (47%), Gaps = 5/90 (5%)
Frame = +1
Query: 311 WYFDSGASNHVTHQTNKFQDLTE-----HHGKNSLVVGNGDKLEIVATCSSKLKSLNLDD 365
W DSG +NH+T+ F +L E +N + K + + LK ++ +
Sbjct: 73 WLIDSGCTNHMTYDRELFTELDEVVFSKVKIRNEAYIDVKGKETVAI*GHTGLKLIS--N 246
Query: 366 VLYVPNITKNLLSVSKLAADNNIFVEFDKN 395
VLYV I++NLLSV +L + DKN
Sbjct: 247 VLYVSEISQNLLSVPQLLKKGYKVLFEDKN 336
>CF922226
Length = 667
Score = 36.6 bits (83), Expect = 0.026
Identities = 51/225 (22%), Positives = 77/225 (33%), Gaps = 12/225 (5%)
Frame = -3
Query: 121 IYLKSEFHSIRKGEMK-MEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIV 179
+Y K +S + E + + + L L L+ I + D + L L Y+
Sbjct: 641 LYXKQSLYSFKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYLPKSYSHFK 462
Query: 180 VKL---SDHTTLSWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNN 236
L D +L V R E+ ++ + L A K D +
Sbjct: 461 ETLLFGRDSVSLDEVQTALNSKELNERKEKKSSASGEGLTARGKTFKKDSEFDKKKQKPE 282
Query: 237 NWRGSNFRGWRGGRGRGRSSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADS------ 290
N + G G K C C K HT C R KN +N DS
Sbjct: 281 NQKN----------GEGNIFKIRCYHCKKEGHTRKVCPER-QKNGGSNNRKKDSGNAAIV 135
Query: 291 --DKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTE 333
D S A + S+ + E W DSG S H+T + F+ ++
Sbjct: 134 QDDGYESAEALMVSEKNPET-KWIMDSGCSWHMTPNKSWFEQFSD 3
>NP004897 gag-protease polyprotein
Length = 1923
Score = 34.3 bits (77), Expect = 0.13
Identities = 28/110 (25%), Positives = 41/110 (36%), Gaps = 11/110 (10%)
Frame = +1
Query: 252 RGRSSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQG-----------SHNAFI 300
+ + K C CGK H C+H + + S+ K H +
Sbjct: 1489 KSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSSSRRKMMWVPKHKIVSLVVHTSLR 1668
Query: 301 ASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEI 350
AS DWY DSG S H+T +F E + + G+G K +I
Sbjct: 1669 ASAKE----DWYLDSGCSRHMT-GVKEFLVNIEPCSTSYVTFGDGSKGKI 1803
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 32.3 bits (72), Expect = 0.48
Identities = 16/38 (42%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Frame = +2
Query: 240 GSNFRGWRG-GRGRGRSSKAP-CQVCGKTNHTAINCFH 275
G + G G GRG GR P C CG+ H A +C+H
Sbjct: 359 GGRYGGGEGRGRGFGRRGGGPECYNCGRIGHLARDCYH 472
>CK605939
Length = 458
Score = 32.0 bits (71), Expect = 0.63
Identities = 16/39 (41%), Positives = 23/39 (58%)
Frame = +3
Query: 217 ATANVANKFDHRDNRFNSNNNWRGSNFRGWRGGRGRGRS 255
A A+++++ R R ++ WRGS WRGGRG G S
Sbjct: 138 AGASLSSRASLRRPREKGDDGWRGS---AWRGGRGAGTS 245
>TC215566 similar to UP|Q9FSG4 (Q9FSG4) Wound-induced GSK-3-like protein
(Fragment), partial (62%)
Length = 1401
Score = 32.0 bits (71), Expect = 0.63
Identities = 30/138 (21%), Positives = 53/138 (37%), Gaps = 4/138 (2%)
Frame = +3
Query: 58 SSDSSKSNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTR 117
SSD +N +++ + N++ T Q +TSK+ + S R
Sbjct: 408 SSDPGGDSNSKRAKFEPETEGKADEKTNTIETICTDQEQPIDTSKETSNVGTSDVSTVAR 587
Query: 118 SQIIYLKSEFHSIRK--GEMKMEDYLIKMKNLAD--KLKLAGNPISNSDLIIQTLNGLDS 173
++ KS F + K EMK+ D K N D ++GN +I + G D
Sbjct: 588 TE----KSGFEELPKELNEMKIRDEKSKNNNEKDIEATVVSGNGTETGQIITTDIGGRDG 755
Query: 174 EYNPIVVKLSDHTTLSWV 191
+ + + +H W+
Sbjct: 756 QPKQTISYMDEHVEWDWI 809
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.130 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,538,918
Number of Sequences: 63676
Number of extensions: 269791
Number of successful extensions: 1860
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1823
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1848
length of query: 482
length of database: 12,639,632
effective HSP length: 101
effective length of query: 381
effective length of database: 6,208,356
effective search space: 2365383636
effective search space used: 2365383636
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144765.7


