

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.20 + phase: 0
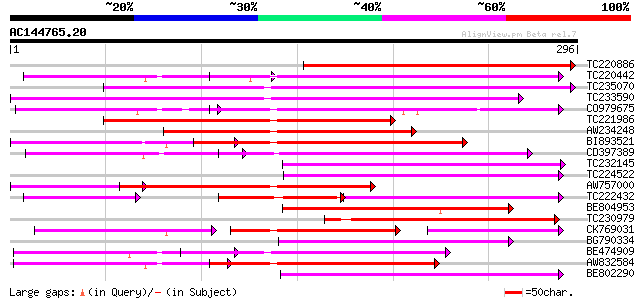
(296 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220886 weakly similar to UP|Q9FNN7 (Q9FNN7) Dbj|BAA90805.1, pa... 233 8e-62
TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 139 2e-33
TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 138 3e-33
TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-... 137 4e-33
CO979675 133 1e-31
TC221986 weakly similar to GB|BAC16401.1|23237826|AP003749 selen... 125 2e-29
AW234248 125 2e-29
BI893521 125 2e-29
CD397389 similar to GP|6691205|gb| F1K23.11 {Arabidopsis thalian... 117 6e-27
TC232145 116 1e-26
TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, pa... 115 3e-26
AW757000 similar to PIR|C86440|C86 PPR-repeat protein [imported]... 113 1e-25
TC222432 77 1e-25
BE804953 similar to GP|9758973|dbj selenium-binding protein-like... 111 3e-25
TC230979 weakly similar to UP|Q6Z8F8 (Q6Z8F8) Selenium-binding p... 107 6e-24
CK769031 84 1e-23
BG790334 weakly similar to GP|2160154|gb| F19K23.18 gene product... 106 1e-23
BE474909 103 1e-22
AW832584 102 3e-22
BE802290 100 1e-21
>TC220886 weakly similar to UP|Q9FNN7 (Q9FNN7) Dbj|BAA90805.1, partial (11%)
Length = 852
Score = 233 bits (594), Expect = 8e-62
Identities = 111/142 (78%), Positives = 126/142 (88%)
Frame = +3
Query: 154 ACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSV 213
ACSHKGLVDKARW FDHM K+YKI+PD KHYGC+VDLLSR+GLLEEA+Q+I TAP QNS
Sbjct: 6 ACSHKGLVDKARWNFDHMAKQYKILPDIKHYGCIVDLLSRFGLLEEAHQMIKTAPLQNSA 185
Query: 214 VLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEM 273
+LWRTLLGACRTQ N ELA++SF+QLAKL +L DGDYVLLSNIYAEA RWDEVER+RSEM
Sbjct: 186 ILWRTLLGACRTQGNVELAKVSFQQLAKLGRLTDGDYVLLSNIYAEAERWDEVERVRSEM 365
Query: 274 DYLHVPRQAGYSQINMKESDRL 295
LHVP+Q YSQI+M ESD+L
Sbjct: 366 IGLHVPKQVAYSQIDMTESDKL 431
>TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (25%)
Length = 706
Score = 139 bits (350), Expect = 2e-33
Identities = 71/185 (38%), Positives = 108/185 (58%)
Frame = +1
Query: 105 KTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKA 164
K V W A++ GLA+HG E FT+M+++ I P+ +TF +L ACSH GL ++
Sbjct: 4 KCVCAWTAIIGGLAIHGKGREALDWFTQMQKA---GINPNSITFTAILTACSHAGLTEEG 174
Query: 165 RWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACR 224
+ F+ M Y I P +HYGCMVDL+ R GLL+EA + I + P + + +W LL AC+
Sbjct: 175 KSLFESMSSVYNIKPSMEHYGCMVDLMGRAGLLKEAREFIESMPVKPNAAIWGALLNACQ 354
Query: 225 TQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGY 284
+ EL + K L +L G Y+ L++IYA AG W++V R+RS++ + + G
Sbjct: 355 LHKHFELGKEIGKILIELDPDHSGRYIHLASIYAAAGEWNQVVRVRSQIKHRGLLNHPGC 534
Query: 285 SQINM 289
S I +
Sbjct: 535 SSITL 549
Score = 57.0 bits (136), Expect = 1e-08
Identities = 37/139 (26%), Positives = 67/139 (47%), Gaps = 7/139 (5%)
Frame = +1
Query: 8 SWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESL 67
+W ++I G AL+ F++MQ AG+ P +T +IL AC+ G E G ++ES+
Sbjct: 16 AWTAIIGGLAIHGKGREALDWFTQMQKAGINPNSITFTAILTACSHAGLTEEGKSLFESM 195
Query: 68 KV---CEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMK-TVSCWNAMVIGLAVHGYC 123
+ +E Y +V++ + G L A E M +K + W A++ +H +
Sbjct: 196 SSVYNIKPSMEHY--GCMVDLMGRAGLLKEAREFIESMPVKPNAAIWGALLNACQLHKHF 369
Query: 124 E---EVFQLFTEMEESLGG 139
E E+ ++ E++ G
Sbjct: 370 ELGKEIGKILIELDPDHSG 426
>TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (22%)
Length = 875
Score = 138 bits (348), Expect = 3e-33
Identities = 83/246 (33%), Positives = 125/246 (50%)
Frame = +2
Query: 50 ACAETGALEIGHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSC 109
AC+ + G ++ L ++ Y+G ALV+ Y KCG+L+ A F + V+
Sbjct: 2 ACSCLCSFRQGQLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNVAA 181
Query: 110 WNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFD 169
W A++ G A HG E LF M L I P+ TF+GVL AC+H GLV + F
Sbjct: 182 WTALINGYAYHGLGSEAILLFRSM---LHQGIVPNAATFVGVLSACNHAGLVCEGLRIFH 352
Query: 170 HMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNT 229
M + Y + P +HY C+VDLL R G L+EA + I+ P + ++W LL A +
Sbjct: 353 SMQRCYGVTPTIEHYTCVVDLLGRSGHLKEAEEFIIKMPIEADGIIWGALLNASWFWKDM 532
Query: 230 ELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
E+ E + ++L L +V+LSN+YA GRW + +LR + L + + G S I +
Sbjct: 533 EVGERAAEKLFSLDPNPIFAFVVLSNMYAILGRWGQKTKLRKRLQSLELRKDPGCSWIEL 712
Query: 290 KESDRL 295
L
Sbjct: 713 NNKIHL 730
>TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-containing
protein-like, partial (5%)
Length = 1094
Score = 137 bits (346), Expect = 4e-33
Identities = 86/269 (31%), Positives = 135/269 (49%), Gaps = 1/269 (0%)
Frame = +2
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MP+ ++ SW S+I G AL +F +M V+P + TL + L ACA +L+ G
Sbjct: 59 MPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQFTLSTCLFACATIASLKHG 238
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKT-VSCWNAMVIGLAV 119
+I+ L + K + + A+VNMY KCG+L A +FN + K V WN M++ LA
Sbjct: 239 RQIHAFLVLNNIKPNTIVVCAIVNMYSKCGSLETARRVFNFIGNKQDVVLWNTMILALAH 418
Query: 120 HGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVP 179
+GY E + M L ++P++ TF+G+L AC H GLV + F M + +VP
Sbjct: 419 YGYGIEAIMMLYNM---LKIGVKPNKGTFVGILNACCHSGLVQEGLQLFKSMTSEHGVVP 589
Query: 180 DSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQL 239
D +HY + +LL + E+ + + + + + +G CR N + L
Sbjct: 590 DQEHYTRLANLLGQARCFNESVKDLQMMDCKPGDHVCNSSIGVCRMHGNIDHGAEVAAFL 769
Query: 240 AKLKQLIDGDYVLLSNIYAEAGRWDEVER 268
KL+ Y LLS YA G+W+ VE+
Sbjct: 770 IKLQPQSSAAYELLSRTYAALGKWELVEK 856
>CO979675
Length = 732
Score = 133 bits (334), Expect = 1e-31
Identities = 75/190 (39%), Positives = 110/190 (57%), Gaps = 5/190 (2%)
Frame = -2
Query: 105 KTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKA 164
+ + CWNAM+ GLA+HG ++F+EME++ I+PD +TFI V ACS+ G+ +
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKT---GIKPDDITFIAVFTACSYSGMAHEG 561
Query: 165 RWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQII--MTAPFQN---SVVLWRTL 219
D M Y+I P S+HYGC+VDLLSR GL EA +I +T+ N + WR
Sbjct: 560 LQLLDKMSSLYEIEPKSEHYGCLVDLLSRAGLFGEAMVMIRRITSTSWNGSEETLAWRAF 381
Query: 220 LGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVP 279
L AC +LAE + K+L +L+ G YVLLSN+YA +G+ + R+R+ M V
Sbjct: 380 LSACCNHGQAQLAERAAKRLLRLEN-HSGVYVLLSNLYAASGKHSDARRVRNMMRNKGVD 204
Query: 280 RQAGYSQINM 289
+ G S + +
Sbjct: 203 KAPGCSSVEI 174
Score = 60.5 bits (145), Expect = 9e-10
Identities = 38/111 (34%), Positives = 60/111 (53%), Gaps = 3/111 (2%)
Frame = -2
Query: 4 RNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKI 63
R+ V WN+MI+G D A AL++FSEM+ G+KP ++T I++ AC+ +G G ++
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAHEGLQL 552
Query: 64 YE---SLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWN 111
+ SL E K E Y LV++ + G L E ++ T + WN
Sbjct: 551 LDKMSSLYEIEPKSEHY--GCLVDLLSRAG---LFGEAMVMIRRITSTSWN 414
>TC221986 weakly similar to GB|BAC16401.1|23237826|AP003749 selenium-binding
protein-like {Oryza sativa (japonica cultivar-group);} ,
partial (10%)
Length = 853
Score = 125 bits (315), Expect = 2e-29
Identities = 61/152 (40%), Positives = 97/152 (63%)
Frame = +1
Query: 50 ACAETGALEIGHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSC 109
AC E G+L++G ++++ ++E +LG AL++MY KCGNL A +F+ M+M+T++
Sbjct: 4 ACTEMGSLKLGRRVHDFALKNGFELEPFLGTALIDMYSKCGNLDDARTVFDMMQMRTLAT 183
Query: 110 WNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFD 169
WN M+ L VHGY +E +F EME++ + PD +TF+GVL AC + ++ A+ YF+
Sbjct: 184 WNTMITSLGVHGYRDEALSIFEEMEKA---NEVPDAITFVGVLSACVYMNDLELAQKYFN 354
Query: 170 HMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAY 201
M Y I P +HY CMV++ +R L+E Y
Sbjct: 355 LMTDHYGITPILEHYTCMVEIHTRAIKLDEIY 450
>AW234248
Length = 399
Score = 125 bits (315), Expect = 2e-29
Identities = 64/132 (48%), Positives = 83/132 (62%)
Frame = +2
Query: 81 ALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGS 140
+LV+MY KCGN+ A +F ++ WNAM++GLA HG EE Q F EM+
Sbjct: 11 SLVDMYAKCGNIEDARGLFKRTNTSRIASWNAMIVGLAQHGNAEEALQFFEEMKSR---G 181
Query: 141 IRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEA 200
+ PDRVTFIGVL ACSH GLV +A F M K Y I P+ +HY C+VD LSR G + EA
Sbjct: 182 VTPDRVTFIGVLSACSHSGLVSEAYENFYSMQKIYGIEPEIEHYSCLVDALSRAGRIREA 361
Query: 201 YQIIMTAPFQNS 212
++I + PF+ S
Sbjct: 362 EKVISSMPFEAS 397
>BI893521
Length = 423
Score = 125 bits (314), Expect = 2e-29
Identities = 61/143 (42%), Positives = 89/143 (61%)
Frame = +1
Query: 97 EIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACS 156
++FN M + V W +++ G A HG+ + +LF EM E ++P+ VT+I VL ACS
Sbjct: 1 QVFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLEI---GVKPNEVTYIAVLSACS 171
Query: 157 HKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLW 216
H GL+D+A +F+ M + I P +HY CMVDLL R GLL EA + I + PF ++W
Sbjct: 172 HVGLIDEAWKHFNSMHYNHSISPRMEHYACMVDLLGRSGLLLEAIEFINSMPFDADALVW 351
Query: 217 RTLLGACRTQSNTELAEISFKQL 239
RT LG+CR NT+L E + K++
Sbjct: 352 RTFLGSCRVHRNTKLGEHAAKKI 420
Score = 59.3 bits (142), Expect = 2e-09
Identities = 40/123 (32%), Positives = 58/123 (46%), Gaps = 3/123 (2%)
Frame = +1
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
M RN ++W S+I+G ALELF EM GVKP EVT I++L AC+ G ++
Sbjct: 16 MGYRNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACSHVGLIDEA 195
Query: 61 HKIYESLKVCEHKIESYLGN--ALVNMYCKCGNLSLAWEIFNGMKMKT-VSCWNAMVIGL 117
K + S+ H I + + +V++ + G L A E N M W +
Sbjct: 196 WKHFNSMHY-NHSISPRMEHYACMVDLLGRSGLLLEAIEFINSMPFDADALVWRTFLGSC 372
Query: 118 AVH 120
VH
Sbjct: 373 RVH 381
>CD397389 similar to GP|6691205|gb| F1K23.11 {Arabidopsis thaliana}, partial
(16%)
Length = 626
Score = 117 bits (293), Expect = 6e-27
Identities = 65/165 (39%), Positives = 89/165 (53%), Gaps = 1/165 (0%)
Frame = -2
Query: 110 WNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFD 169
W +M+ G +G+ +E +LF +M G I P+ VT + L AC+H GLVDK
Sbjct: 625 WTSMMDGYGKNGFPDEALELFVKMPTEYG--IVPNYVTLLSALSACAHAGLVDKGWEIIQ 452
Query: 170 HMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNT 229
M Y + P +HY CMVDLL R G+L +A++ IM P + +W LL +CR N
Sbjct: 451 SMENEYLVKPGMEHYACMVDLLGRAGMLNQAWEFIMRIPEKPISDVWAALLSSCRLHGNI 272
Query: 230 ELAEISFKQLAKLKQL-IDGDYVLLSNIYAEAGRWDEVERLRSEM 273
ELA+++ +L KL G YV LSN AG W+ V LR M
Sbjct: 271 ELAKLAANELFKLNATGRPGAYVALSNTLVAAG*WESVTELREIM 137
Score = 53.5 bits (127), Expect = 1e-07
Identities = 39/121 (32%), Positives = 62/121 (51%), Gaps = 5/121 (4%)
Frame = -2
Query: 9 WNSMIAGCVSVRDYAGALELFSEMQNA-GVKPTEVTLISILGACAETGALEIGHKIYESL 67
W SM+ G ALELF +M G+ P VTL+S L ACA G ++ G +I +S+
Sbjct: 625 WTSMMDGYGKNGFPDEALELFVKMPTEYGIVPNYVTLLSALSACAHAGLVDKGWEIIQSM 446
Query: 68 K---VCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVS-CWNAMVIGLAVHGYC 123
+ + + +E Y +V++ + G L+ AWE + K +S W A++ +HG
Sbjct: 445 ENEYLVKPGMEHYA--CMVDLLGRAGMLNQAWEFIMRIPEKPISDVWAALLSSCRLHGNI 272
Query: 124 E 124
E
Sbjct: 271 E 269
>TC232145
Length = 817
Score = 116 bits (291), Expect = 1e-26
Identities = 61/148 (41%), Positives = 88/148 (59%)
Frame = +3
Query: 143 PDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQ 202
P+ +TF+ VL ACSH G +D YF+ M + I P +HY CMVDLL R G LEEA +
Sbjct: 15 PEYITFVSVLSACSHTGKIDDGFKYFNSMANVHNIKPGLEHYACMVDLLGRVGRLEEACR 194
Query: 203 IIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGR 262
I + PF+ ++W LLGAC +N E+ ++L KL+ G+Y+LLSNIY G
Sbjct: 195 FIESMPFEPDSLVWGALLGACGKHANVEMGREVAERLFKLEPDNPGNYMLLSNIYIRHGM 374
Query: 263 WDEVERLRSEMDYLHVPRQAGYSQINMK 290
+E + +R M V +++G S I++K
Sbjct: 375 LEEADEVRRLMGINGVRKESGCSWIDVK 458
>TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, partial (18%)
Length = 534
Score = 115 bits (287), Expect = 3e-26
Identities = 58/146 (39%), Positives = 85/146 (57%)
Frame = +2
Query: 144 DRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQI 203
D +TF+ +L AC G V+++ F MV Y I P S+HY C+VD++SR G L+ A +I
Sbjct: 2 DGITFLSLLSACCRAGKVNESMNLFSLMVDNYGIPPRSEHYACLVDVMSRAGQLQRACKI 181
Query: 204 IMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGRW 263
I PF+ +W +L AC N EL E++ +++ L G YV+LSNIYA AG+W
Sbjct: 182 INEMPFKADSSIWGAVLAACSVHLNVELGELAARRILNLDPFNSGAYVMLSNIYAAAGKW 361
Query: 264 DEVERLRSEMDYLHVPRQAGYSQINM 289
+V R+R M V +Q YS + +
Sbjct: 362 KDVHRIRVLMKEQGVKKQTAYSWLQI 439
>AW757000 similar to PIR|C86440|C86 PPR-repeat protein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 442
Score = 113 bits (282), Expect = 1e-25
Identities = 54/134 (40%), Positives = 82/134 (60%)
Frame = +3
Query: 58 EIGHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGL 117
E G I+ + K+++ +G AL+ MY KCG + ++EIFNG+K K + W +++ GL
Sbjct: 3 EQGKWIHNYIDENRIKVDAVVGTALIEMYAKCGCIEKSFEIFNGLKEKDTTSWTSIICGL 182
Query: 118 AVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKI 177
A++G E +LF M+ ++PD +TF+ VL ACSH GLV++ R F M Y I
Sbjct: 183 AMNGKPSEALELFKAMQTC---GLKPDDITFVAVLSACSHAGLVEEGRKLFHSMSSMYHI 353
Query: 178 VPDSKHYGCMVDLL 191
P+ +HYGC +DLL
Sbjct: 354 EPNLEHYGCFIDLL 395
Score = 60.5 bits (145), Expect = 9e-10
Identities = 26/72 (36%), Positives = 42/72 (58%)
Frame = +3
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
+ E++ SW S+I G + ALELF MQ G+KP ++T +++L AC+ G +E G
Sbjct: 135 LKEKDTTSWTSIICGLAMNGKPSEALELFKAMQTCGLKPDDITFVAVLSACSHAGLVEEG 314
Query: 61 HKIYESLKVCEH 72
K++ S+ H
Sbjct: 315 RKLFHSMSSMYH 350
>TC222432
Length = 951
Score = 77.4 bits (189), Expect(2) = 1e-25
Identities = 39/115 (33%), Positives = 64/115 (54%)
Frame = +2
Query: 175 YKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEI 234
+ P +HYGCMVDLL R G L+EA +I T P+ + ++ + L AC ++ AE
Sbjct: 200 FGFAPQVEHYGCMVDLLGRAGCLDEAENLIQTMPYDANGIILSSFLFACGYFNDVLRAER 379
Query: 235 SFKQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
K++ K+ + + G+YV+L N+YA RW +VE ++ M ++ S I +
Sbjct: 380 VLKEVVKMDEDVAGNYVMLRNLYATRQRWTDVEDVKQMMKKRGTSKEVACSVIEI 544
Score = 56.6 bits (135), Expect(2) = 1e-25
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 1/67 (1%)
Frame = +3
Query: 110 WNAMVIGLAVHGYCEEVFQLFTEM-EESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYF 168
WNA++ G AV+G +E ++F M EE G P+ VT IGVL AC+H GLV++ R +F
Sbjct: 15 WNALINGFAVNGCAKEALEVFARMIEEGFG----PNEVTMIGVLSACNHCGLVEEGRRWF 182
Query: 169 DHMVKRY 175
+ M +RY
Sbjct: 183 NAM-ERY 200
Score = 44.7 bits (104), Expect = 5e-05
Identities = 22/61 (36%), Positives = 34/61 (55%)
Frame = +3
Query: 8 SWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESL 67
SWN++I G ALE+F+ M G P EVT+I +L AC G +E G + + ++
Sbjct: 12 SWNALINGFAVNGCAKEALEVFARMIEEGFGPNEVTMIGVLSACNHCGLVEEGRRWFNAM 191
Query: 68 K 68
+
Sbjct: 192E 194
>BE804953 similar to GP|9758973|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (12%)
Length = 370
Score = 111 bits (278), Expect = 3e-25
Identities = 56/123 (45%), Positives = 79/123 (63%), Gaps = 2/123 (1%)
Frame = +2
Query: 143 PDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQ 202
PD VTF+GVL ACSH GLVD+ +F M + Y++ P +H+ CMVDLL R G +++ +
Sbjct: 2 PDHVTFVGVLSACSHVGLVDEGFEHFKSMGEVYELAPRIEHFSCMVDLLGRAGDVKKLEE 181
Query: 203 IIMTAPFQNSVVLWRTLLGAC--RTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEA 260
I T P + ++WRT+LGAC NTEL + K L +L+ L +YVLLSN++A
Sbjct: 182 FIKTMPMNPNALIWRTILGACCRANSRNTELGRRAAKMLIELESLNAVNYVLLSNMHAAG 361
Query: 261 GRW 263
G+W
Sbjct: 362 GKW 370
>TC230979 weakly similar to UP|Q6Z8F8 (Q6Z8F8) Selenium-binding protein-like,
partial (61%)
Length = 837
Score = 107 bits (267), Expect = 6e-24
Identities = 51/123 (41%), Positives = 80/123 (64%)
Frame = +2
Query: 165 RWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACR 224
R F+HM+K P +HYGCMVDL+ R G+L+EAY +I + P + + V+WR+LL AC+
Sbjct: 11 RMQFEHMIK-----PTIQHYGCMVDLMGRAGMLKEAYDLIKSMPIKPNDVVWRSLLSACK 175
Query: 225 TQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGY 284
N E+ EI+ + KL + GDY++L+N+YA A +W V R+R+EM ++ + G+
Sbjct: 176 VHHNLEIGEIAADNIFKLNKHNPGDYLVLANMYARAQKWANVARIRTEMVEKNLVQTPGF 355
Query: 285 SQI 287
S +
Sbjct: 356 SLV 364
Score = 33.1 bits (74), Expect = 0.15
Identities = 26/77 (33%), Positives = 40/77 (51%), Gaps = 3/77 (3%)
Frame = +2
Query: 15 GC-VSVRDYAGAL-ELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESL-KVCE 71
GC V + AG L E + +++ +KP +V S+L AC LEIG +++ K+ +
Sbjct: 56 GCMVDLMGRAGMLKEAYDLIKSMPIKPNDVVWRSLLSACKVHHNLEIGEIAADNIFKLNK 235
Query: 72 HKIESYLGNALVNMYCK 88
H YL L NMY +
Sbjct: 236 HNPGDYL--VLANMYAR 280
>CK769031
Length = 827
Score = 84.0 bits (206), Expect(2) = 1e-23
Identities = 43/90 (47%), Positives = 58/90 (63%), Gaps = 1/90 (1%)
Frame = +2
Query: 116 GLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVK-R 174
GLA++GY +E + F ++ ES RP+ VTF+ V AC H GLVD+ R YF+ M
Sbjct: 14 GLAINGYGKEALKRFEDLVES---GARPNEVTFLAVYTACCHNGLVDEGRKYFNEMTSPH 184
Query: 175 YKIVPDSKHYGCMVDLLSRWGLLEEAYQII 204
Y + P +HYGCMVDLL R GL+ EA ++I
Sbjct: 185 YNLSPCLEHYGCMVDLLCRAGLVGEAVELI 274
Score = 43.1 bits (100), Expect(2) = 1e-23
Identities = 25/71 (35%), Positives = 35/71 (49%)
Frame = +1
Query: 219 LLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHV 278
LL AC T N + K L ++ G YVLLSN+YA +W EV +R M +
Sbjct: 298 LLSACNT*GNVGFTQEMLKSLQNVEFQDSGIYVLLSNLYATNKKWAEVRSIRRLMKQKGI 477
Query: 279 PRQAGYSQINM 289
+ G S I++
Sbjct: 478 SKAPGSSIISV 510
Score = 42.0 bits (97), Expect = 3e-04
Identities = 22/98 (22%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Frame = +2
Query: 14 AGCVSVRDYAG-ALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESLKVCEH 72
+G +++ Y AL+ F ++ +G +P EVT +++ AC G ++ G K + + +
Sbjct: 8 SGGLAINGYGKEALKRFEDLVESGARPNEVTFLAVYTACCHNGLVDEGRKYFNEMTSPHY 187
Query: 73 KIESYLGN--ALVNMYCKCGNLSLAWEIFNGMKMKTVS 108
+ L + +V++ C+ G + A E+ + + +S
Sbjct: 188 NLSPCLEHYGCMVDLLCRAGLVGEAVELIKIFRSQELS 301
>BG790334 weakly similar to GP|2160154|gb| F19K23.18 gene product
{Arabidopsis thaliana}, partial (19%)
Length = 390
Score = 106 bits (264), Expect = 1e-23
Identities = 53/123 (43%), Positives = 73/123 (59%)
Frame = +2
Query: 141 IRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEA 200
I P +TFI VL AC+H GLV++ F M+ Y I P +H+ +VD+L R G L+EA
Sbjct: 20 IHPTYITFISVLNACAHAGLVEEGWRQFKSMINDYGIEPRVEHFASLVDILGRQGQLQEA 199
Query: 201 YQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEA 260
+I T PF+ +W LLGACR +N ELA ++ L +L+ YVLL N+YA
Sbjct: 200 MDLINTMPFKPDKAVWGALLGACRVHNNVELALVAADALIRLEPESSAPYVLLYNMYANL 379
Query: 261 GRW 263
G+W
Sbjct: 380 GQW 388
>BE474909
Length = 423
Score = 103 bits (256), Expect = 1e-22
Identities = 53/141 (37%), Positives = 81/141 (56%)
Frame = +2
Query: 90 GNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFI 149
G +S A +F G+ K V WN++++G A HG LF +M L + PD +T
Sbjct: 8 GYVSDAVYVFKGINEKNVVSWNSVIVGCAQHGCGMWALALFNQM---LREGVDPDGITVT 178
Query: 150 GVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPF 209
G+L ACSH G++ KAR +F + ++ + +HY MVD+L R G LEEA ++M+ P
Sbjct: 179 GLLSACSHSGMLQKARCFFRYFGQKRSVTLTIEHYTSMVDVLGRCGELEEAEAVVMSMPM 358
Query: 210 QNSVVLWRTLLGACRTQSNTE 230
+ + ++W LL ACR SN +
Sbjct: 359 KANSMVWLALLSACRKHSNLD 421
Score = 60.1 bits (144), Expect = 1e-09
Identities = 39/122 (31%), Positives = 60/122 (48%), Gaps = 4/122 (3%)
Frame = +2
Query: 3 ERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGH- 61
E+N VSWNS+I GC AL LF++M GV P +T+ +L AC+ +G L+
Sbjct: 50 EKNVVSWNSVIVGCAQHGCGMWALALFNQMLREGVDPDGITVTGLLSACSHSGMLQKARC 229
Query: 62 --KIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVS-CWNAMVIGLA 118
+ + + IE Y ++V++ +CG L A + M MK S W A++
Sbjct: 230 FFRYFGQKRSVTLTIEHY--TSMVDVLGRCGELEEAEAVVMSMPMKANSMVWLALLSACR 403
Query: 119 VH 120
H
Sbjct: 404 KH 409
>AW832584
Length = 373
Score = 102 bits (253), Expect = 3e-22
Identities = 52/120 (43%), Positives = 73/120 (60%)
Frame = +2
Query: 105 KTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKA 164
K V WN+M+ G A+HG+ E+ +LF+ M PD TF G+L AC+H GLV++
Sbjct: 8 KDVVSWNSMIQGFAMHGHGEKALELFSRMVPE---GFEPDTYTFDGLLCACTHAGLVNEG 178
Query: 165 RWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACR 224
R YF M K Y IVP +HYG M+DLL R G L+EA+ ++ P + + ++ TLL A R
Sbjct: 179 RMYFYSMEKVYGIVPQVEHYGYMMDLLGRGGHLKEAFTLLRGMPMEPNAIILGTLLNASR 358
Score = 50.1 bits (118), Expect = 1e-06
Identities = 35/117 (29%), Positives = 58/117 (48%), Gaps = 3/117 (2%)
Frame = +2
Query: 3 ERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHK 62
+++ VSWNSMI G ALELFS M G +P T +L AC G + G
Sbjct: 5 KKDVVSWNSMIQGFAMHGHGEKALELFSRMVPEGFEPDTYTFDGLLCACTHAGLVNEGRM 184
Query: 63 IYESLKV---CEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIG 116
+ S++ ++E Y ++++ + G+L A+ + GM M+ NA+++G
Sbjct: 185 YFYSMEKVYGIVPQVEHY--GYMMDLLGRGGHLKEAFTLLRGMPMEP----NAIILG 337
>BE802290
Length = 481
Score = 99.8 bits (247), Expect = 1e-21
Identities = 50/148 (33%), Positives = 83/148 (55%)
Frame = -1
Query: 142 RPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAY 201
+P+ + F+ + ACSH L ++ F+ M Y I P +HYGCMVDL+ GLL+EA
Sbjct: 454 KPNSIPFMAISPACSHAALTEEGTSSFESMSSVYNIKPSMEHYGCMVDLMGGAGLLKEAR 275
Query: 202 QIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAG 261
+ + + P + + +W LL AC+ EL + + K +L G Y+ L++IYA A
Sbjct: 274 EFVESMPVKPNAAIWGALLNACQLHKYFELGKETGKIQIELDPDHSGRYIHLASIYAAAR 95
Query: 262 RWDEVERLRSEMDYLHVPRQAGYSQINM 289
W++V R+RS++ + + +G S I +
Sbjct: 94 EWNQVTRVRSQIKHRGLLNYSGCSSITL 11
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,697,183
Number of Sequences: 63676
Number of extensions: 191169
Number of successful extensions: 985
Number of sequences better than 10.0: 125
Number of HSP's better than 10.0 without gapping: 897
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 941
length of query: 296
length of database: 12,639,632
effective HSP length: 97
effective length of query: 199
effective length of database: 6,463,060
effective search space: 1286148940
effective search space used: 1286148940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144765.20


