

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.10 - phase: 0
(552 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
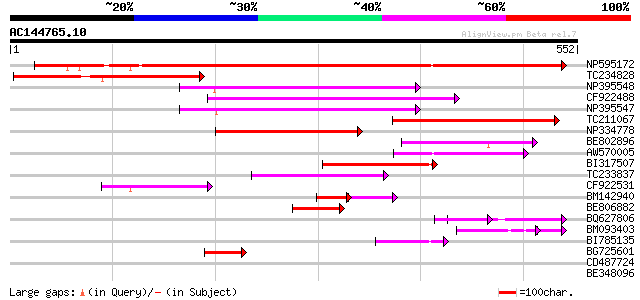
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 408 e-114
TC234828 183 2e-46
NP395548 reverse transcriptase [Glycine max] 169 3e-42
CF922488 155 3e-38
NP395547 reverse transcriptase [Glycine max] 152 5e-37
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 142 4e-34
NP334778 reverse transcriptase [Glycine max] 109 3e-24
BE802896 105 4e-23
AW570005 100 1e-21
BI317507 98 1e-20
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 92 6e-19
CF922531 69 7e-12
BM142940 40 1e-08
BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, pa... 53 4e-07
BQ627806 48 1e-05
BM093403 43 7e-05
BI785135 45 8e-05
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 44 2e-04
CD487724 42 0.001
BE348096 40 0.003
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 408 bits (1049), Expect = e-114
Identities = 222/543 (40%), Positives = 333/543 (60%), Gaps = 25/543 (4%)
Frame = +1
Query: 25 PLSMFGRDFEVDLVCLPLLGMDVIFGMNWL--------EYNRVCINCFNK----TVHFSS 72
PL + G++ +V + L + G DVI G WL +Y + + F T+
Sbjct: 1357 PLHIQGQEVKVPVYLLQISGADVILGSTWLATLGPHVADYAALTLKFFQNDKFITLQGEG 1536
Query: 73 AEEESGAQFLTTKQLKQLERDGILMFSLMASLSLENQVVIDRLP------------VVNE 120
E + AQ ++L+ + + A ++ +V D L +++
Sbjct: 1537 NSEATQAQLHHFRRLQNTKS----IEECFAIQLIQKEVPEDTLKDLPTNIDPELAILLHT 1704
Query: 121 FHEVFPDEIP-DVPPEREVEFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFV 179
+ +VF +P +PP+RE + +I L G+ V + PY ++ +++K ++++L + +
Sbjct: 1705 YAQVFA--VPASLPPQREQDHAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGII 1878
Query: 180 RPSVSPWGAPVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKI 239
+PS SP+ P+LLVKKKDGS R C DYR LN +T+K+ +P+P +D+L+D+L GA+ FSK+
Sbjct: 1879 QPSNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKL 2058
Query: 240 DLRSGYHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFV 299
DLRSGYHQI V+ ED +KTAFRT +GHYE+ VMPFG+TNAP F MN+IF L KFV
Sbjct: 2059 DLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFV 2238
Query: 300 VVFIDDILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIA 359
+VF DDILIYS ++H KHL+ VLQ LK+ +L+A+LSKC F +EV +LGH SG G++
Sbjct: 2239 LVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVS 2418
Query: 360 VDPSKVDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQ 419
++ +KV AV W TP +V ++R FLGL GYYRRFI+ ++ +A PLT L K SF+W+ +
Sbjct: 2419 MENTKVQAVLDWPTPNNVKQLRGFLGLTGYYRRFIKSYANIAGPLTDLLQK-DSFLWNNE 2595
Query: 420 CESSFNELKQRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHE 479
E++F +LK+ +T AP+L LP +PF++ DAS G+G VL Q+G +AY S++L
Sbjct: 2596 AEAAFVKLKKAMTEAPVLSLPDFSQPFILETDASGIGVGAVLGQNGHPIAYFSKKLAPRM 2775
Query: 480 KNSPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKD 539
+ + EL A+ L +RHYL G++F + +D +SLK L DQ Q+ WL
Sbjct: 2776 QKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSLKSLMDQSLQTPEQQAWLHKFLG 2955
Query: 540 YDF 542
YDF
Sbjct: 2956 YDF 2964
>TC234828
Length = 857
Score = 183 bits (464), Expect = 2e-46
Identities = 92/190 (48%), Positives = 127/190 (66%), Gaps = 4/190 (2%)
Frame = +3
Query: 4 EMVVETPAKGSVTTSLVCLRCPLSMFGRDFEVDLVCLPLLGMDVIFGMNWLEYNRVCINC 63
+MVV TP VTTS VCL+CP+ + GR F DL+CLPL +DVI GM+WL N + ++C
Sbjct: 306 DMVVSTPTSEPVTTSRVCLKCPIIVEGRSFMADLICLPLAHLDVILGMDWLSTNHIFLDC 485
Query: 64 FNKTVHFSSAEEESGAQFLTTKQLKQ----LERDGILMFSLMASLSLENQVVIDRLPVVN 119
K + F G + ++ LK+ E + + + ++ S+ +E + +PVV+
Sbjct: 486 KEKMLVF-------GGDVVPSEPLKEDAANEETEDVRTYMVLFSMYVEEDAEVSCIPVVS 644
Query: 120 EFHEVFPDEIPDVPPEREVEFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFV 179
EF EVFPD++ ++PPEREVEF ID+VPGA VS+APY MS ELAE+K Q++DLL K+FV
Sbjct: 645 EFPEVFPDDVCELPPEREVEFIIDVVPGANPVSIAPYRMSPVELAEVKAQVQDLLSKQFV 824
Query: 180 RPSVSPWGAP 189
RPS SPWGAP
Sbjct: 825 RPSASPWGAP 854
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 169 bits (428), Expect = 3e-42
Identities = 96/254 (37%), Positives = 143/254 (55%), Gaps = 19/254 (7%)
Frame = +1
Query: 166 LKKQLEDLLDKKFVRP-SVSPWGAPVLLVKKKDG------------------SMRLCIDY 206
++K++ LL+ + P S S W +PVL+V KK+G S +LCIDY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 207 RQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYGH 266
R+LN+ T K+ +PLP +D ++++L G + +D GY+QI V +D +K AF +G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 267 YEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKHLKIVLQI 326
+ Y+ +PFG+ NAP F M IF ++K + VF+DD ++ E K L++VLQ
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 327 LKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTEIRSFLGL 386
E L KC F + E LGH S GI VD +K+D + + P +V IRSFLG
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 387 AGYYRRFIEGFSKL 400
A +YRRFI+ F+K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>CF922488
Length = 741
Score = 155 bits (393), Expect = 3e-38
Identities = 85/246 (34%), Positives = 134/246 (53%)
Frame = +3
Query: 193 VKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKD 252
V K+DG + +C+DYR LN + K+++PLP I+ L+D FS +D SGY+QIK+
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 253 EDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKI 312
EDM+KT F T +G + YK M FG+ N + M +F + K + V++DD+++ S+
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 313 EEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWE 372
EEEH +L+ + + L++ +L +KC F + L + S GI VD +KV + +
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 373 TPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLT 432
P + +++ FLG Y RFI PL L K + WD C +F +KQ L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 433 TAPILI 438
+L+
Sbjct: 723 NPHVLV 740
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 152 bits (383), Expect = 5e-37
Identities = 90/254 (35%), Positives = 133/254 (51%), Gaps = 19/254 (7%)
Frame = +1
Query: 166 LKKQLEDLLDKKFVRP-SVSPWGAPVLLVKKKDGSM------------------RLCIDY 206
++K++ LL+ + P S S W +PV +V KK G R+CIDY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 207 RQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYGH 266
R+LN+ T K+ YPLP +D ++ +L + +D SGY+QI V +D +KTAF +
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 267 YEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKHLKIVLQI 326
+ Y+ MPFG+ NA F M IF ++K + VF+DD + +L+ VLQ
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 327 LKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTEIRSFLGL 386
++ L KC F + E LGH S GI V K+D + + P +V I SFLG
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 387 AGYYRRFIEGFSKL 400
G+YRRFI+ F+K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 142 bits (358), Expect = 4e-34
Identities = 72/163 (44%), Positives = 101/163 (61%)
Frame = +1
Query: 373 TPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLT 432
T KSV +IRSF GLA +YRRF+ FS +A PL +L K +F W + E +F LK++LT
Sbjct: 100 TLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLT 279
Query: 433 TAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAV 492
AP+L LP + F + CDAS G+ VL+Q G +AY S +L N PT+D EL A+
Sbjct: 280 KAPVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYAL 459
Query: 493 VFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLE 535
+ + W H+L F + SDH+SLKY+ + +LN R +W+E
Sbjct: 460 IRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWVE 588
Score = 40.4 bits (93), Expect = 0.002
Identities = 24/76 (31%), Positives = 37/76 (48%)
Frame = +2
Query: 341 FWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKL 400
F + + F G V NG+ +DP K+ A+ +W P V EI LG + + IEG +
Sbjct: 2 FCVDNIFFSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEI---LGASMG*QASIEGSFLI 172
Query: 401 ALPLTQLTYKGKSFVW 416
+L L L+ +W
Sbjct: 173 SLQLHHLSMSW*RRIW 220
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 109 bits (273), Expect = 3e-24
Identities = 55/143 (38%), Positives = 87/143 (60%)
Frame = +3
Query: 201 RLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTAF 260
R+C+DYR LN+ + K+ +PLP ID LM + +FS +D SGY+QIK+ EDM+KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 261 RTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKHL 320
T +G + YKVM FG+ N + M +F + K + ++D+++ S++EEEH +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 321 KIVLQILKERKLYAKLSKCEFWL 343
+ + L++ +L KC F L
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>BE802896
Length = 416
Score = 105 bits (263), Expect = 4e-23
Identities = 60/137 (43%), Positives = 83/137 (59%), Gaps = 4/137 (2%)
Frame = -2
Query: 382 SFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILILPK 441
SFLG AG+YRRFI F K+ALPL+ L K F ++ +C+ +F+ LK+ L T PI+ P
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPD 236
Query: 442 PEEPFVVYCDASKFGLGGVLMQD----GKVVAYASRQLRVHEKNSPTHDLELAAVVFVLK 497
PF + CDAS + LG VL Q +V+ Y+SR L + N T + EL A+VF L+
Sbjct: 235 WTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALE 56
Query: 498 IWRHYLYGSRFEVFSDH 514
+ YL G+R V+ DH
Sbjct: 55 KFHSYLLGTRIIVYIDH 5
>AW570005
Length = 413
Score = 100 bits (250), Expect = 1e-21
Identities = 54/132 (40%), Positives = 78/132 (58%)
Frame = -2
Query: 374 PKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTT 433
P++ +R FL L G+YRRFI+G++ +A PL+ L K SFVW + + +F LK +T
Sbjct: 406 PRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKD-SFVWSPEADVAFQALKNVVTN 230
Query: 434 APILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVV 493
+L LP +PF V DAS +G VL Q+G +A+ S++ S T+ ELAA+
Sbjct: 229 TLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAIT 50
Query: 494 FVLKIWRHYLYG 505
V+K WR YL G
Sbjct: 49 NVVKKWRQYLLG 14
>BI317507
Length = 359
Score = 97.8 bits (242), Expect = 1e-20
Identities = 52/113 (46%), Positives = 71/113 (62%), Gaps = 1/113 (0%)
Frame = -1
Query: 305 DILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSK 364
+ILIYS + H HL VL +LK+ +L A KC F + + +LGHV S + +A+D +K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 365 VDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLA-LPLTQLTYKGKSFVW 416
V +V +W PK+V + SFL L GYYR+FI+ + KLA PLT LT K F W
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLT-KNDGFKW 18
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 92.0 bits (227), Expect = 6e-19
Identities = 48/133 (36%), Positives = 76/133 (57%)
Frame = +2
Query: 236 FSKIDLRSGYHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFL 295
FS +D SGY+QI + ED++KT F T +G + Y+VM FG+ N + M +FH +
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 296 DKFVVVFIDDILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSG 355
K + V++DD++ S+ E EH +L + L++ +L +KC F + LG + S
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 356 NGIAVDPSKVDAV 368
GI +DP KV A+
Sbjct: 362 KGIEIDPEKVKAL 400
>CF922531
Length = 602
Score = 68.6 bits (166), Expect = 7e-12
Identities = 44/114 (38%), Positives = 66/114 (57%), Gaps = 6/114 (5%)
Frame = -2
Query: 90 LERDGILMFSLMASLSLENQVVIDRLP-----VVNEFHEVFPDEIP-DVPPEREVEFSID 143
L++ L+ S SLS I+ LP +++EF ++FP EIP +PP R +E ID
Sbjct: 343 LKQSFYLLLSRETSLSTVTIPTIETLPPKVQELLHEFGDIFPKEIPLGLPPLRGIEHQID 164
Query: 144 LVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPWGAPVLLVKKKD 197
LVP A L + Y + E E++ Q+++LL+K +V+ S+S VLLV KKD
Sbjct: 163 LVPRASLPNRPTYRTNPQETKEIESQVKELLEKGWVQESLSLCVVLVLLVPKKD 2
>BM142940
Length = 422
Score = 40.0 bits (92), Expect(2) = 1e-08
Identities = 21/34 (61%), Positives = 23/34 (66%)
Frame = -1
Query: 299 VVVFIDDILIYSKIEEEHAKHLKIVLQILKERKL 332
+ F DILIYS E +H KHLKIVLQ KE KL
Sbjct: 260 ICFFSYDILIYSINEGKHEKHLKIVLQTFKEAKL 159
Score = 37.7 bits (86), Expect(2) = 1e-08
Identities = 15/47 (31%), Positives = 25/47 (52%)
Frame = -3
Query: 331 KLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSV 377
K+ A KC F + +L H+ S G++ DP K++ + W PK +
Sbjct: 165 KVGANRKKCSFGCKKFEYLVHMISVKGVSADPKKLEVMVAWPIPKEM 25
>BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, partial (3%)
Length = 153
Score = 52.8 bits (125), Expect = 4e-07
Identities = 25/51 (49%), Positives = 33/51 (64%)
Frame = -1
Query: 276 VTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKHLKIVLQI 326
+TNAP F MN IF L K+V+VF DDIL+YS EH HL++V ++
Sbjct: 153 LTNAPTSFQCLMNHIFQHALRKYVLVFFDDILVYSSTWHEHLCHLEVVFKV 1
>BQ627806
Length = 435
Score = 48.1 bits (113), Expect = 1e-05
Identities = 34/116 (29%), Positives = 53/116 (45%)
Frame = +1
Query: 427 LKQRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHD 486
+K R ILILP + + F ++ + + S+ LR + T+
Sbjct: 76 VKLRFWVYQILILPLSLKQMLRELVWGPFFHRTIIPLPFSFMPFCSKLLR-----ASTYV 240
Query: 487 LELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDF 542
ELAA+ +K WR YL G F + +DH+SLK L Q Q+ +L L +D+
Sbjct: 241 RELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQTPEQQIYLARLMGFDY 408
Score = 45.4 bits (106), Expect = 6e-05
Identities = 22/57 (38%), Positives = 34/57 (59%)
Frame = +3
Query: 414 FVWDAQCESSFNELKQRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAY 470
F W+ + + +F++LK L AP+L LP FVV DAS G+G +L Q+ +A+
Sbjct: 21 FHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHPLAF 191
>BM093403
Length = 419
Score = 42.7 bits (99), Expect(2) = 7e-05
Identities = 25/81 (30%), Positives = 44/81 (53%)
Frame = -1
Query: 436 ILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFV 495
+L+ P +PF+V DA G+G ++MQ+ + +AY S+ L + N EL A+V
Sbjct: 404 VLVTPNFSKPFIVEADALGSGIGAIVMQESRPIAYYSKALLICPYNIVMQ--ELLALVMP 231
Query: 496 LKIWRHYLYGSRFEVFSDHKS 516
++ W ++ + VF+ KS
Sbjct: 230 IQYWCP-IFWAENSVFTTKKS 171
Score = 21.9 bits (45), Expect(2) = 7e-05
Identities = 10/28 (35%), Positives = 14/28 (49%)
Frame = -2
Query: 515 KSLKYLFDQKELNMRQRRWLELLKDYDF 542
KSL +L +Q Q+ W + YDF
Sbjct: 178 KSLHHLLEQ*ITTQNQQDWFVKILRYDF 95
>BI785135
Length = 420
Score = 45.1 bits (105), Expect = 8e-05
Identities = 22/71 (30%), Positives = 36/71 (49%)
Frame = +1
Query: 357 GIAVDPSKVDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVW 416
G+ ++P ++ + +W TP S+ I F L +Y+RF+ FS L PL +L + W
Sbjct: 208 GVPMNPKRIKVIPEWPTPPSIR*IWGFHNLTNFYKRFVP*FSILVAPLIELV-RNHVPSW 384
Query: 417 DAQCESSFNEL 427
+ E F L
Sbjct: 385 EEAQEMGFQTL 417
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 43.5 bits (101), Expect = 2e-04
Identities = 20/41 (48%), Positives = 26/41 (62%)
Frame = -3
Query: 190 VLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQL 230
V++VKK +G R+C DY LN K+ YPLP ID + D L
Sbjct: 124 VVMVKKPNGKWRICTDYIDLN*ACPKDAYPLPNIDHMTDGL 2
>CD487724
Length = 676
Score = 41.6 bits (96), Expect = 0.001
Identities = 37/154 (24%), Positives = 70/154 (45%), Gaps = 17/154 (11%)
Frame = +3
Query: 20 VCLRCPLSMFGRDFEVDLVCLPLLGMDVIFGMNWL--------EYNRVCINCF--NKTVH 69
+C P+S+ +F V L LP++G +++ G+ WL +YN + + F ++ V
Sbjct: 162 ICEAIPISIQNIEFLVHLYVLPIVGANIVLGVQWLKTLGPILVDYNSLSMQFFYQHRLVQ 341
Query: 70 FSSAEEESGAQFLTTKQLKQLERDGILMFSLMASLSLENQVVIDRLP-------VVNEFH 122
E E+ L QL++L + + ++ EN P ++++F
Sbjct: 342 L-KGESEAQLGLLNHHQLRRLHQTHEPVTYFHIAILTENTSPTSSPPLPQPIQHLLDQFS 518
Query: 123 EVFPDEIPDVPPEREVEFSIDLVPGAKLVSMAPY 156
+F +PP RE + I L+P ++ V+M Y
Sbjct: 519 ALFQ*P-QGLPPARETDHHIHLLP*SEPVNMRLY 617
>BE348096
Length = 445
Score = 40.0 bits (92), Expect = 0.003
Identities = 17/39 (43%), Positives = 23/39 (58%)
Frame = -2
Query: 465 GKVVAYASRQLRVHEKNSPTHDLELAAVVFVLKIWRHYL 503
G+V Y ++ H + PTHDLEL A+V WRHY+
Sbjct: 183 GQVATYVLP*VKTH*RIYPTHDLELVAMVCAFTFWRHYI 67
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,024,625
Number of Sequences: 63676
Number of extensions: 322063
Number of successful extensions: 1688
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 1673
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1682
length of query: 552
length of database: 12,639,632
effective HSP length: 102
effective length of query: 450
effective length of database: 6,144,680
effective search space: 2765106000
effective search space used: 2765106000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144765.10


