

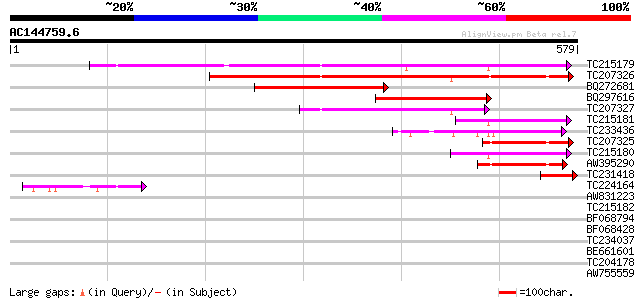
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144759.6 + phase: 0
(579 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC215179 homologue to UP|Q94IR2 (Q94IR2) Carotenoid 9,10-9',10' ... 333 1e-91
TC207326 homologue to UP|Q9FS24 (Q9FS24) Neoxanthin cleavage enz... 293 1e-79
BQ272681 similar to GP|22335707|db nine-cis-epoxycarotenoid diox... 218 6e-57
BQ297616 similar to GP|2828292|emb neoxanthin cleavage enzyme-li... 183 2e-46
TC207327 homologue to UP|Q9M6E8 (Q9M6E8) 9-cis-epoxycarotenoid d... 144 1e-34
TC215181 homologue to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoi... 90 2e-18
TC233436 weakly similar to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycaro... 90 3e-18
TC207325 homologue to UP|Q9FS24 (Q9FS24) Neoxanthin cleavage enz... 89 4e-18
TC215180 homologue to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoi... 86 3e-17
AW395290 similar to GP|22335703|dbj nine-cis-epoxycarotenoid dio... 80 2e-15
TC231418 similar to UP|Q8LP14 (Q8LP14) Nine-cis-epoxycarotenoid ... 60 3e-09
TC224164 similar to UP|Q9M6E8 (Q9M6E8) 9-cis-epoxycarotenoid dio... 53 4e-07
AW831223 homologue to GP|22335695|dbj nine-cis-epoxycarotenoid d... 39 0.005
TC215182 homologue to UP|Q94IR2 (Q94IR2) Carotenoid 9,10-9',10' ... 39 0.008
BF068794 34 0.16
BF068428 weakly similar to GP|13876587|gb| carotenoid 9 10-9' 10... 32 1.0
TC234037 weakly similar to PIR|T06608|T06608 disease resistance ... 31 1.7
BE661601 weakly similar to GP|12483839|gb| p6.9 {Heliocoverpa ar... 30 3.9
TC204178 homologue to UP|TBB7_MAIZE (Q41784) Tubulin beta-7 chai... 29 5.1
AW755559 29 5.1
>TC215179 homologue to UP|Q94IR2 (Q94IR2) Carotenoid 9,10-9',10' cleavage
dioxygenase, complete
Length = 1934
Score = 333 bits (854), Expect = 1e-91
Identities = 194/513 (37%), Positives = 284/513 (54%), Gaps = 21/513 (4%)
Frame = +1
Query: 82 DIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQ 141
D++ + + S P H L+ NFAPV E PPT+ +KG LP LNG ++R GPNP+
Sbjct: 199 DLLEKLVVKFLYDSSLPHHYLTGNFAPV-SETPPTKDLPVKGYLPDCLNGEFVRVGPNPK 375
Query: 142 FLPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNV--FSGFNS 199
F P YH FDGDGM+H ++I +GKAT SR+V+T + K E G + G
Sbjct: 376 FAPVAGYHWFDGDGMIHGLRIKDGKATYVSRFVRTSRLKQEEYFGGSKFMKIGDLKGLFG 555
Query: 200 LIGLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDI 259
L+ + +VL Y G G NT+L +L AL E+D PY IKV +GD+
Sbjct: 556 LLMVNIHMLRTKWKVLDASY----GTGTANTALVYHHGKLLALSEADKPYAIKVFEDGDL 723
Query: 260 QTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGVKHNDVPVFS 319
QT+G D++ +L + TAHPK+D TGE F + Y P++TY +G H+ VP+ +
Sbjct: 724 QTLGMLDYDKRLGHSFTAHPKVDPFTGEMFTFGYAHTPPYITYRVISKDGYMHDPVPI-T 900
Query: 320 MKRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRS-DPSKVPRIGILPRYADNES 378
+ P +HDFAIT+ YA+F D+ L P +M+ + + S D +K R G+LPRYA +E
Sbjct: 901 VSDPIMMHDFAITENYAIFLDLPLIFRPKEMVKNKTLIFSFDSTKKARFGVLPRYAKDEK 1080
Query: 379 KMKWFDVPGLNIMHLINAWDEEDGK---TVTIVAPNI-LCVEHMMERLELIHEMIEKVRI 434
++WF++P I H NAW+EED T + PN+ L E+LE + ++R
Sbjct: 1081LIRWFELPNCFIFHNANAWEEEDEVVLITCRLQNPNLDLVGGTAKEKLENFSNELYEMRF 1260
Query: 435 NVETGIVTRQPLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDV------- 487
N++TG +++ LSA +D +N + G+K R++Y + + K +G++K D+
Sbjct: 1261NMKTGEASQKKLSASAVDFPRVNESYTGRKQRYVYGTTLDSIAKVTGIIKFDLHAEPDNG 1440
Query: 488 -----LKGEEVGCRLYGEDCYGGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESRFLVM 540
+ G G G YG E +V R G EEDDGYL+ +VHDE G+S V+
Sbjct: 1441KTKLEVGGNVQGLYDLGPGKYGSEAVYVPRVPGTDSEEDDGYLICFVHDENTGKSFVHVI 1620
Query: 541 DAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDI 573
+AK+ + VA V+LP RVPYGFH FV E +
Sbjct: 1621NAKTMSADPVAVVELPHRVPYGFHAFFVTEEQL 1719
>TC207326 homologue to UP|Q9FS24 (Q9FS24) Neoxanthin cleavage enzyme, partial
(63%)
Length = 1509
Score = 293 bits (751), Expect = 1e-79
Identities = 152/377 (40%), Positives = 231/377 (60%), Gaps = 6/377 (1%)
Frame = +2
Query: 205 ARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGR 264
AR + AR L G + +G+G+ N L F N L A+ E DLPY +++TPNGD+ T+GR
Sbjct: 44 ARLLLFYARSLFGLVDGSHGMGVANAGLVYFNNHLLAMSEDDLPYHLRITPNGDLTTVGR 223
Query: 265 YDFNGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRP 323
Y+FNG+L M AHPK+D T A Y ++ +P+L YFRF +GVK DV + +K P
Sbjct: 224 YNFNGQLKSTMIAHPKLDPVTNNLHALSYDVVQKPYLKYFRFSPDGVKSPDVEI-PLKEP 400
Query: 324 SFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWF 383
+ +HDFAIT+ + + D Q+ +MI GGSPV D +KV R GIL + A + + MKW
Sbjct: 401 TMMHDFAITENFVVVPDQQVVFKLSEMITGGSPVVYDKNKVSRFGILDKNAKDANDMKWI 580
Query: 384 DVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIEKVRINVETGIVT 442
D P HL NAW+E + + ++ + + + E E + ++ ++R+N++TG T
Sbjct: 581 DAPECFCFHLWNAWEEPENDEIVVIGSCMTPADSIFNECEESLKSILSEIRLNLKTGKST 760
Query: 443 RQPLSAR----NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLY 498
R+P+ + NL+ ++N LG+K +F Y A+ P PK SG K+D+ G EV +Y
Sbjct: 761 RKPIISESEQVNLEAGMVNRNKLGRKTKFGYLALAEPWPKVSGFAKVDLFSG-EVKKYMY 937
Query: 499 GEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRR 558
GE+ +GGEP F+ +EDDG+++++VHDEK+ +S +++AK+ + E A VKLP R
Sbjct: 938 GEERFGGEPLFLPNGVDGDEDDGHILAFVHDEKEWKSELQIVNAKTLKLE--ASVKLPSR 1111
Query: 559 VPYGFHGLFVKESDITR 575
VPYGFHG F+ D+ +
Sbjct: 1112VPYGFHGTFIHSKDLRK 1162
>BQ272681 similar to GP|22335707|db nine-cis-epoxycarotenoid dioxygenase4
{Pisum sativum}, partial (23%)
Length = 421
Score = 218 bits (555), Expect = 6e-57
Identities = 102/137 (74%), Positives = 116/137 (84%)
Frame = +3
Query: 251 IKVTPNGDIQTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGV 310
+ VTP+GDI+T+GR+DF+GKL +MTAHPKID DT ECFA+RYG + PFLTYFRFD NG
Sbjct: 6 VNVTPDGDIETLGRHDFDGKLTFSMTAHPKIDPDTAECFAFRYGPVPPFLTYFRFDGNGK 185
Query: 311 KHNDVPVFSMKRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGIL 370
KH DVP+FSM PSFLHDFAITKKYA+F DIQL +NPLDMI GGSPV S SKVPRIGIL
Sbjct: 186 KHEDVPIFSMLTPSFLHDFAITKKYAIFCDIQLGMNPLDMISGGSPVGSVASKVPRIGIL 365
Query: 371 PRYADNESKMKWFDVPG 387
PR A +ES MKWF+VPG
Sbjct: 366 PRDAKDESMMKWFEVPG 416
>BQ297616 similar to GP|2828292|emb neoxanthin cleavage enzyme-like protein
{Arabidopsis thaliana}, partial (19%)
Length = 431
Score = 183 bits (464), Expect = 2e-46
Identities = 81/119 (68%), Positives = 105/119 (88%)
Frame = -2
Query: 374 ADNESKMKWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMMERLELIHEMIEKVR 433
A +ES MKWF+VPG NI+H INAW+E++G+TV +VAPNIL +EH +ER+EL+H M+EK+R
Sbjct: 370 AKDESMMKWFEVPGFNIIHAINAWEEDEGRTVVLVAPNILSMEHALERMELVHAMVEKIR 191
Query: 434 INVETGIVTRQPLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEE 492
I ++TGI+TRQP+SARNLD AVIN F+GKK+RF+YAA+G+PMPK SGVVK+DV KGEE
Sbjct: 190 IELDTGIITRQPVSARNLDFAVINPAFVGKKNRFVYAAVGDPMPKISGVVKLDVSKGEE 14
>TC207327 homologue to UP|Q9M6E8 (Q9M6E8) 9-cis-epoxycarotenoid dioxygenase,
partial (32%)
Length = 599
Score = 144 bits (363), Expect = 1e-34
Identities = 73/199 (36%), Positives = 115/199 (57%), Gaps = 5/199 (2%)
Frame = +1
Query: 297 RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSP 356
+P+L YFRF NGVK DV + +K P+ +HDFAIT+ + + D Q+ +MI GGSP
Sbjct: 4 KPYLKYFRFSPNGVKSPDVEI-PLKEPTMMHDFAITENFVVIPDQQVVFKLSEMISGGSP 180
Query: 357 VRSDPSKVPRIGILPRYADNESKMKWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVE 416
V D +KV R GIL + A + + MKW D P HL NAW+E + + ++ + +
Sbjct: 181 VVYDKNKVSRFGILNKNAKDPNGMKWIDAPECFCFHLWNAWEEPENDEIVVIGSCMTPAD 360
Query: 417 HMMERL-ELIHEMIEKVRINVETGIVTRQPLSAR----NLDLAVINNEFLGKKHRFIYAA 471
+ E + ++ ++R+N++TG TR+P+ + NL+ ++N LG+K +F Y A
Sbjct: 361 SIFNECDESLKSVLSEIRLNLKTGKSTRKPIISESQQVNLEAGMVNRNKLGRKTQFAYLA 540
Query: 472 IGNPMPKFSGVVKIDVLKG 490
+ P PK SG K+D+ G
Sbjct: 541 LAEPWPKVSGFAKVDLFSG 597
>TC215181 homologue to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoid
dioxygenase1, partial (25%)
Length = 741
Score = 90.1 bits (222), Expect = 2e-18
Identities = 49/132 (37%), Positives = 71/132 (53%), Gaps = 14/132 (10%)
Frame = +2
Query: 456 INNEFLGKKHRFIYAAIGNPMPKFSGVVKIDV------------LKGEEVGCRLYGEDCY 503
+N + G+K R++Y + + K +G++K D+ + G G G Y
Sbjct: 155 VNESYTGRKQRYVYGTTLDSIAKVTGIIKFDLHAEPDNGKTKLEVGGNVQGLYDLGPGKY 334
Query: 504 GGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPY 561
G E +V R G EEDDGYL+ +VHDE G+S V++AK+ + VA V+LP RVPY
Sbjct: 335 GSEAVYVPRVPGTDSEEDDGYLICFVHDENTGKSFVHVINAKTMSADPVAVVELPHRVPY 514
Query: 562 GFHGLFVKESDI 573
GFH FV E +
Sbjct: 515 GFHAFFVTEEQL 550
>TC233436 weakly similar to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoid
dioxygenase1, partial (9%)
Length = 806
Score = 89.7 bits (221), Expect = 3e-18
Identities = 62/210 (29%), Positives = 103/210 (48%), Gaps = 33/210 (15%)
Frame = +1
Query: 392 HLINAWDEEDGKTVTI--------VAPNILCVEHMMERLELIHEMIEKVRINVETGIVTR 443
H+IN++ EDG V + + P E L HE R+N++TG V
Sbjct: 22 HIINSF--EDGHEVVVRGCRSLDSLIPGPDPSLKEFEWLSRCHEW----RLNMQTGEVKE 183
Query: 444 QPLSARNL---DLAVINNEFLGKKHRFIYAAIGNPM-------PKFSGVVKIDV------ 487
+ L N+ D +IN F+G ++R+ Y + +P+ PK+ G+ K+
Sbjct: 184 RGLCGANIVYMDFPMINGNFIGIRNRYAYTQVVDPIASSTQDVPKYGGLAKLYFEESCAK 363
Query: 488 --LKGEE-------VGCRLYGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFL 538
++ E V C ++ ++ + FV R+ G EEDDG+++++VH+E
Sbjct: 364 FSMRNREQPEEPIRVECHMFEKNTFCSGAAFVPRDGGLEEDDGWIIAFVHNEDTNICEVH 543
Query: 539 VMDAKSPEFEIVAEVKLPRRVPYGFHGLFV 568
++D K E VA++ +PRRVPYGFHG F+
Sbjct: 544 IIDTKKFSGETVAKITMPRRVPYGFHGAFM 633
>TC207325 homologue to UP|Q9FS24 (Q9FS24) Neoxanthin cleavage enzyme, partial
(14%)
Length = 608
Score = 89.4 bits (220), Expect = 4e-18
Identities = 42/92 (45%), Positives = 64/92 (68%)
Frame = +2
Query: 484 KIDVLKGEEVGCRLYGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAK 543
K+D+ GE V +YGE+ +GGEP F+ +EDDGY++++VHDEK+ +S +++AK
Sbjct: 5 KVDLFSGE-VNKYMYGEERFGGEPLFLPNGVDGDEDDGYILAFVHDEKEWKSELQIVNAK 181
Query: 544 SPEFEIVAEVKLPRRVPYGFHGLFVKESDITR 575
+ + E A VKLP RVPYGFHG F+ +D+ +
Sbjct: 182 TLKLE--ASVKLPSRVPYGFHGTFIHSNDLRK 271
>TC215180 homologue to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoid
dioxygenase1, partial (26%)
Length = 708
Score = 86.3 bits (212), Expect = 3e-17
Identities = 48/137 (35%), Positives = 71/137 (51%), Gaps = 14/137 (10%)
Frame = +1
Query: 451 LDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDV------------LKGEEVGCRLY 498
+D +N + G+K R++Y + + K +G++K D+ + G G
Sbjct: 1 VDFPRVNESYTGRKQRYVYGTTLDSIAKVTGIIKFDLHAEPDHGKTKLEVGGNVQGLYDL 180
Query: 499 GEDCYGGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLP 556
G YG E +V R G EEDDGYL+ + HDE +S V++AK+ + VA V+LP
Sbjct: 181 GPGKYGSEAVYVPRVPGTDSEEDDGYLIFFAHDENTRKSFVHVINAKTMSADPVAVVELP 360
Query: 557 RRVPYGFHGLFVKESDI 573
RVPYGFH FV E +
Sbjct: 361 HRVPYGFHAFFVTEEQL 411
>AW395290 similar to GP|22335703|dbj nine-cis-epoxycarotenoid dioxygenase3
{Pisum sativum}, partial (14%)
Length = 396
Score = 80.5 bits (197), Expect = 2e-15
Identities = 44/93 (47%), Positives = 59/93 (63%), Gaps = 1/93 (1%)
Frame = +1
Query: 478 KFSGVVKIDVLKGEEVGCRLYGEDCYGGEPFFVAREDGE-EEDDGYLVSYVHDEKKGESR 536
K SGV K+D+ GE V YGE +GGEPFF+ G ED+GY++++VHDE +S
Sbjct: 13 KVSGVAKVDLESGE-VKRHEYGERRFGGEPFFLPTRGGNGNEDEGYVMAFVHDEMTWQSE 189
Query: 537 FLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVK 569
+++A + E A V LP RVPYGFHG FV+
Sbjct: 190 LQILNALDLKLE--ATVMLPSRVPYGFHGTFVE 282
>TC231418 similar to UP|Q8LP14 (Q8LP14) Nine-cis-epoxycarotenoid
dioxygenase4, partial (6%)
Length = 490
Score = 60.1 bits (144), Expect = 3e-09
Identities = 25/37 (67%), Positives = 34/37 (91%)
Frame = +1
Query: 543 KSPEFEIVAEVKLPRRVPYGFHGLFVKESDITRLSVS 579
+SPE ++VA V+LPRRVPYGFHGLFVKES++ ++S+S
Sbjct: 13 RSPELDVVAAVRLPRRVPYGFHGLFVKESELRKVSLS 123
>TC224164 similar to UP|Q9M6E8 (Q9M6E8) 9-cis-epoxycarotenoid dioxygenase,
partial (22%)
Length = 557
Score = 52.8 bits (125), Expect = 4e-07
Identities = 47/152 (30%), Positives = 68/152 (43%), Gaps = 26/152 (17%)
Frame = +2
Query: 14 PPTTYIPQFN------------TASVRTKEKPQTSQPK------TLRTTP---TKTLLAL 52
PPTT +F T S++T P+ QP T TTP TK ++A
Sbjct: 122 PPTTLKKRFPCNSNSNSNSNAITCSLQTLHFPKKYQPTSTTTTTTTTTTPARETKPVIAS 301
Query: 53 LTTTTQKLKNTNKTLQTSSISALILNTLDDIINTFI-----DPPIKPSVDPRHVLSQNFA 107
+ T L + LQ ++ SAL D++ T + P+ + DPR ++ NFA
Sbjct: 302 PSETKHPLPQSWNFLQKAAASAL------DMVETALVSHERKHPLPKTADPRVQIAGNFA 463
Query: 108 PVLDELPPTQCKVIKGTLPPSLNGVYIRNGPN 139
PV E P + G +P + GVY+RNG N
Sbjct: 464 PV-PEHPAQHSLPVAGKIPKCIEGVYVRNGAN 556
>AW831223 homologue to GP|22335695|dbj nine-cis-epoxycarotenoid dioxygenase1
{Pisum sativum}, partial (8%)
Length = 419
Score = 39.3 bits (90), Expect = 0.005
Identities = 15/33 (45%), Positives = 22/33 (66%)
Frame = +2
Query: 360 DPSKVPRIGILPRYADNESKMKWFDVPGLNIMH 392
D +K R G+LPRYA +E ++WF++P I H
Sbjct: 179 DSTKKARFGVLPRYAKDEKLIRWFELPNCFIFH 277
>TC215182 homologue to UP|Q94IR2 (Q94IR2) Carotenoid 9,10-9',10' cleavage
dioxygenase, partial (6%)
Length = 546
Score = 38.5 bits (88), Expect = 0.008
Identities = 18/36 (50%), Positives = 24/36 (66%)
Frame = +1
Query: 539 VMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDIT 574
V++AK+ + VA V+ P RVPYGFH FV E +T
Sbjct: 1 VINAKTMSADPVAVVEXPHRVPYGFHAFFVTEVCVT 108
>BF068794
Length = 383
Score = 34.3 bits (77), Expect = 0.16
Identities = 24/71 (33%), Positives = 35/71 (48%), Gaps = 6/71 (8%)
Frame = +1
Query: 19 IPQFNTASVRTKEKPQTSQPKTLRTTPTKTLLA------LLTTTTQKLKNTNKTLQTSSI 72
+PQF+ S T P TS +T T+PT TL + +TT + + T + +SS
Sbjct: 25 LPQFSHNSPSTTTTPTTSSWRTSWTSPTMTLSSPMPPFDSITTDSSTVTTTVHSCNSSSF 204
Query: 73 SALILNTLDDI 83
S NT+ DI
Sbjct: 205SGSDPNTVPDI 237
>BF068428 weakly similar to GP|13876587|gb| carotenoid 9 10-9' 10' cleavage
dioxygenase {Phaseolus vulgaris}, partial (14%)
Length = 241
Score = 31.6 bits (70), Expect = 1.0
Identities = 18/43 (41%), Positives = 21/43 (47%)
Frame = +2
Query: 100 HVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQF 142
H S NF PV D PP V KG LP L ++ + P P F
Sbjct: 116 HYFSSNFTPVSDTSPPNDLPV-KGYLPDCLIVQFVISQPYPIF 241
>TC234037 weakly similar to PIR|T06608|T06608 disease resistance protein
homolog F16J13.80 - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (3%)
Length = 858
Score = 30.8 bits (68), Expect = 1.7
Identities = 23/68 (33%), Positives = 32/68 (46%), Gaps = 2/68 (2%)
Frame = +1
Query: 73 SALILNTLDDIINTFIDPPIKPSVDPRHV--LSQNFAPVLDELPPTQCKVIKGTLPPSLN 130
S +L LD N F++PP + ++ + LS N P L LP LPP+L
Sbjct: 259 SLSLLQDLDLSGNNFVNPPAQCIINLSMLQNLSFNDCPRLKSLP---------MLPPNLQ 411
Query: 131 GVYIRNGP 138
G+Y N P
Sbjct: 412 GLYANNCP 435
>BE661601 weakly similar to GP|12483839|gb| p6.9 {Heliocoverpa armigera
nucleopolyhedrovirus G4}, partial (56%)
Length = 658
Score = 29.6 bits (65), Expect = 3.9
Identities = 23/87 (26%), Positives = 38/87 (43%), Gaps = 1/87 (1%)
Frame = +1
Query: 10 ACKQPPTTYIPQFNTASVRTKEKPQTSQPKTLRTTPTKTL-LALLTTTTQKLKNTNKTLQ 68
A PP P ++ T EK +T+ T PT L A TTTT + + +++
Sbjct: 232 AAPAPPAPAAPSPISSPATTPEK------QTVPTNPTPRLGFATTTTTTTRKRRRSRSS* 393
Query: 69 TSSISALILNTLDDIINTFIDPPIKPS 95
S + + T +I+ PP++ S
Sbjct: 394 ISRYCSAAIGTQPEILEPSGTPPLRSS 474
>TC204178 homologue to UP|TBB7_MAIZE (Q41784) Tubulin beta-7 chain (Beta-7
tubulin), complete
Length = 1720
Score = 29.3 bits (64), Expect = 5.1
Identities = 12/22 (54%), Positives = 15/22 (67%), Gaps = 1/22 (4%)
Frame = -1
Query: 100 HVLSQNFAPV-LDELPPTQCKV 120
H+ SQNFAP+ L PP CK+
Sbjct: 220 HITSQNFAPIWLPHCPPWMCKI 155
>AW755559
Length = 435
Score = 29.3 bits (64), Expect = 5.1
Identities = 13/41 (31%), Positives = 22/41 (52%)
Frame = -3
Query: 515 GEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKL 555
G+ + G + ++ +GE L D+K PEFE+ E+ L
Sbjct: 241 GDGFNSGLVEQVTVEDSRGELAMLARDSKGPEFEVHLELAL 119
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.140 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,002,048
Number of Sequences: 63676
Number of extensions: 352518
Number of successful extensions: 1630
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 1586
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1605
length of query: 579
length of database: 12,639,632
effective HSP length: 102
effective length of query: 477
effective length of database: 6,144,680
effective search space: 2931012360
effective search space used: 2931012360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144759.6


