

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144731.8 - phase: 0
(517 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
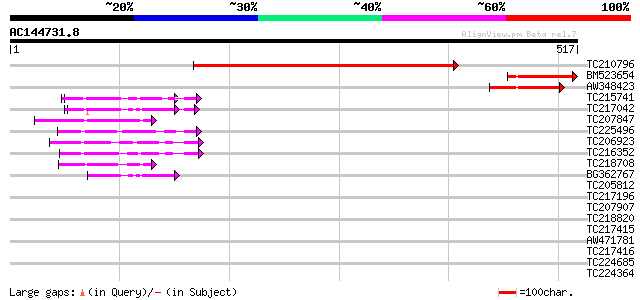
Score E
Sequences producing significant alignments: (bits) Value
TC210796 444 e-125
BM523654 117 1e-26
AW348423 114 1e-25
TC215741 UP|Q6I686 (Q6I686) Protein disufide isomerase-like prot... 60 2e-09
TC217042 UP|Q6I684 (Q6I684) Protein disulfide isomerase-like pro... 59 7e-09
TC207847 similar to UP|Q9LJU2 (Q9LJU2) Emb|CAB38838.1 (AT3g20560... 52 5e-07
TC225496 UP|Q6I687 (Q6I687) Protein disulfide isomerase-like pro... 52 8e-07
TC206923 similar to UP|Q8LAM5 (Q8LAM5) Protein disulfide isomera... 49 4e-06
TC216352 protein disulfide isomerase-like protein [Glycine max] 46 5e-05
TC218708 similar to GB|AAF40463.1|7211992|F13M7 Strong simialrit... 45 1e-04
BG362767 43 3e-04
TC205812 similar to UP|Q9SEU5 (Q9SEU5) Thioredoxin x, partial (64%) 40 0.002
TC217196 similar to GB|CAD83013.1|45771904|ATH551263 mitochondri... 39 0.006
TC207907 similar to UP|Q96313 (Q96313) AT.I.24-13 protein, parti... 37 0.016
TC218820 weakly similar to UP|Q8LGB3 (Q8LGB3) Contains similarit... 36 0.048
TC217415 similar to UP|O23166 (O23166) Thiol-disulfide interchan... 36 0.048
AW471781 35 0.11
TC217416 similar to UP|O23166 (O23166) Thiol-disulfide interchan... 35 0.11
TC224685 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4,... 34 0.14
TC224364 34 0.14
>TC210796
Length = 728
Score = 444 bits (1143), Expect = e-125
Identities = 209/242 (86%), Positives = 223/242 (91%)
Frame = +1
Query: 168 LLNWINKQMSSSFGLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIK 227
LLNWINKQ+ SSFGLDDQKFQNE LSSNVSDP QIARAIYDVEEATS AFDIILE+KMIK
Sbjct: 1 LLNWINKQLGSSFGLDDQKFQNELLSSNVSDPGQIARAIYDVEEATSTAFDIILEHKMIK 180
Query: 228 PETRASLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQI 287
P TR SL+KFLQ+L AHHPSRRCRKG + LVSF DLYPTDF S++KQEDDKSSV+N +I
Sbjct: 181 PGTRTSLIKFLQLLAAHHPSRRCRKGTAEFLVSFDDLYPTDFGSTNKQEDDKSSVRNLKI 360
Query: 288 CGKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVC 347
CGK+VPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRI+DGESQFTFNAIC+FVHNFF+C
Sbjct: 361 CGKDVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRIDDGESQFTFNAICEFVHNFFIC 540
Query: 348 EECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPT 407
EECRQHFY MCSSVS+PFNKARDF LWLWSSHNKVNERL KEEASLGT DP FPKTIWP
Sbjct: 541 EECRQHFYKMCSSVSSPFNKARDFALWLWSSHNKVNERLMKEEASLGTADPIFPKTIWPP 720
Query: 408 KQ 409
KQ
Sbjct: 721 KQ 726
>BM523654
Length = 447
Score = 117 bits (293), Expect = 1e-26
Identities = 57/63 (90%), Positives = 61/63 (96%)
Frame = +1
Query: 455 GNDGTKGAALEDLIVGTNAVVVPVGAALAIAVASCAFGALACYWRSQQKSRKYFHHLHSL 514
GNDG++GA +EDLIV TNA+VVPVGAALAIAVASCAFGALACYWRSQQKSRKYFHHLHSL
Sbjct: 4 GNDGSEGA-VEDLIVATNAIVVPVGAALAIAVASCAFGALACYWRSQQKSRKYFHHLHSL 180
Query: 515 KNI 517
KNI
Sbjct: 181 KNI 189
>AW348423
Length = 511
Score = 114 bits (284), Expect = 1e-25
Identities = 56/69 (81%), Positives = 62/69 (89%)
Frame = -3
Query: 438 NYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVGAALAIAVASCAFGALACY 497
+YY KTL SLYK+ I GNDG++GA +EDLIV TNA+VVPVGAALAIAVASCAFGALACY
Sbjct: 509 DYYSKTLASLYKDNSIVGNDGSEGA-VEDLIVATNAIVVPVGAALAIAVASCAFGALACY 333
Query: 498 WRSQQKSRK 506
WRSQQKSRK
Sbjct: 332 WRSQQKSRK 306
>TC215741 UP|Q6I686 (Q6I686) Protein disufide isomerase-like protein,
complete
Length = 1703
Score = 60.5 bits (145), Expect = 2e-09
Identities = 41/128 (32%), Positives = 61/128 (47%)
Frame = +3
Query: 48 DYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGII 107
D V L+ NF+ + A+VEF+A WC C+ P YEK+ F +V
Sbjct: 195 DDVVVLSEDNFEKEVGQDRG--ALVEFYAPWCGHCKKLAPEYEKLGSSFKKAKSV----- 353
Query: 108 LLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADR 167
L+ +VDC + LC K+ V YP + W P GS EPK+ + RTA+
Sbjct: 354 LIGKVDCDE--HKSLCSKYGVSGYPTIQWFPK-----GSLEPKK-------YEGPRTAES 491
Query: 168 LLNWINKQ 175
L+ ++N +
Sbjct: 492 LVEFVNTE 515
Score = 42.4 bits (98), Expect = 5e-04
Identities = 32/105 (30%), Positives = 49/105 (46%)
Frame = +3
Query: 51 VELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLT 110
V L NF+ V+ D +VEF+A WC C++ P YEKVA F + V ++
Sbjct: 558 VVLTPENFNEVVLDETKD-VLVEFYAPWCGHCKSLAPTYEKVATAFKLEEDV-----VIA 719
Query: 111 RVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSD 155
+D A K L +K+ V +P L + P G + ++ D
Sbjct: 720 NLD-ADKYR-DLAEKYDVSGFPTLKFFPKGNKAGEDYGGGRDLDD 848
>TC217042 UP|Q6I684 (Q6I684) Protein disulfide isomerase-like protein,
complete
Length = 1422
Score = 58.5 bits (140), Expect = 7e-09
Identities = 41/130 (31%), Positives = 62/130 (47%), Gaps = 9/130 (6%)
Frame = +1
Query: 53 LNATNFDAVLKDTPATF---------AVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVH 103
L++ + D V+ T TF A+VEF+A WC C+ P YE++ F +V
Sbjct: 175 LSSASADDVVALTEETFENEVGKDRAALVEFYAPWCGHCKRLAPEYEQLGASFKKTKSV- 351
Query: 104 PGIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDAR 163
L+ +VDC ++ C K+ V YP + W P GS EPK+ + AR
Sbjct: 352 ----LIAKVDCDEHKSV--CGKYGVSGYPTIQWFPK-----GSLEPKK-------YEGAR 477
Query: 164 TADRLLNWIN 173
TA+ L ++N
Sbjct: 478 TAEALAAFVN 507
Score = 49.3 bits (116), Expect = 4e-06
Identities = 36/105 (34%), Positives = 51/105 (48%)
Frame = +1
Query: 51 VELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLT 110
V L+ NFD V+ D +VEF+A WC C+ P YEKVA FN V ++
Sbjct: 556 VVLSPNNFDEVVFDETKD-VLVEFYAPWCGHCKALAPIYEKVAAAFNLDKDV-----VIA 717
Query: 111 RVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSD 155
VD A K L +K+ V YP L + P G +++ ++ D
Sbjct: 718 NVD-ADKYK-DLAEKYGVSGYPTLKFFPKSNKAGENYDGGRDLDD 846
>TC207847 similar to UP|Q9LJU2 (Q9LJU2) Emb|CAB38838.1 (AT3g20560/K10D20_9),
partial (57%)
Length = 1454
Score = 52.4 bits (124), Expect = 5e-07
Identities = 32/113 (28%), Positives = 52/113 (45%), Gaps = 1/113 (0%)
Frame = +1
Query: 23 SSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPAC 82
+ + F ++ V +D+ + + L NFD + P T V F+A WC C
Sbjct: 499 TGAEFHSGTVANAVKHDDEVDEESVEGSFSLTTHNFDKYVHQFPIT--AVNFYAPWCSWC 672
Query: 83 RNYKPHYEKVAKLFNGP-DAVHPGIILLTRVDCASKINIKLCDKFSVGHYPML 134
+ KP +EK AK+ D G I+L +VDC + + LC + + YP +
Sbjct: 673 QRLKPSWEKTAKIMKERYDPEMDGRIILAKVDCTQEGD--LCRRNHIQGYPSI 825
>TC225496 UP|Q6I687 (Q6I687) Protein disulfide isomerase-like protein,
complete
Length = 2048
Score = 51.6 bits (122), Expect = 8e-07
Identities = 36/133 (27%), Positives = 63/133 (47%), Gaps = 1/133 (0%)
Frame = +1
Query: 44 TTDHDYAVELNATNF-DAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAV 102
+++ ++ + L+ +NF D V K F VVEF+A WC C+ P YEK A + +
Sbjct: 175 SSEKEFVLTLDHSNFHDTVSKHD---FIVVEFYAPWCGHCKKLAPEYEKAASILSS---- 333
Query: 103 HPGIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDA 162
H ++L ++D + N L ++ V YP + K + + QE
Sbjct: 334 HDPPVVLAKIDANEEKNKDLASQYDVRGYPTI------KILRNGGKNVQE------YKGP 477
Query: 163 RTADRLLNWINKQ 175
R AD +++++ KQ
Sbjct: 478 READGIVDYLKKQ 516
>TC206923 similar to UP|Q8LAM5 (Q8LAM5) Protein disulfide isomerase-like,
partial (53%)
Length = 1422
Score = 49.3 bits (116), Expect = 4e-06
Identities = 36/140 (25%), Positives = 62/140 (43%)
Frame = +1
Query: 37 HDNDQTGTTDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLF 96
H ++ D V L NF V+++ F +VEF+A WC C+ P Y A
Sbjct: 385 HSTEEAFEVDDKDVVVLKERNFTTVVENN--RFIMVEFYAPWCGHCQALAPEYAAAA--- 549
Query: 97 NGPDAVHPGIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDI 156
+ P ++L +VD + + +L +++ V +P +F+ FV G +P
Sbjct: 550 ---TELKPDGVVLAKVD--ATVENELANEYDVQGFPTVFF-----FVDGVHKP------- 678
Query: 157 HVIDDARTADRLLNWINKQM 176
RT D ++ WI K++
Sbjct: 679 --YTGQRTKDAIVTWIKKKI 732
>TC216352 protein disulfide isomerase-like protein [Glycine max]
Length = 2032
Score = 45.8 bits (107), Expect = 5e-05
Identities = 35/131 (26%), Positives = 55/131 (41%)
Frame = +1
Query: 46 DHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPG 105
D V L NF +K F +VEF+A WC C+ P Y A G D
Sbjct: 340 DEKDVVILKEKNFTDTVKSN--RFVMVEFYAPWCGHCQALAPEYAAAATELKGED----- 498
Query: 106 IILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTA 165
++L +VD + +L ++ V +P + + FV G +P + RT
Sbjct: 499 -VILAKVDATEE--NELAQQYDVQGFPTVHF-----FVDGIHKP---------YNGQRTK 627
Query: 166 DRLLNWINKQM 176
D ++ WI K++
Sbjct: 628 DAIMTWIKKKI 660
>TC218708 similar to GB|AAF40463.1|7211992|F13M7 Strong simialrity to the
disulfide isomerase precursor homolog T21L14.14
gi|2702281 from A. thaliana on BAC gb|AC003033.
{Arabidopsis thaliana;} , partial (77%)
Length = 1274
Score = 44.7 bits (104), Expect = 1e-04
Identities = 27/90 (30%), Positives = 46/90 (51%)
Frame = +2
Query: 45 TDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHP 104
T+ +VELN+ NFD ++ + + +VEFFA WC C+ P ++K + G
Sbjct: 212 TETSSSVELNSGNFDELVIKSKELW-IVEFFAPWCGHCKKLAPEWKKASNNLKGK----- 373
Query: 105 GIILLTRVDCASKINIKLCDKFSVGHYPML 134
+ L VDC ++ + L +F V +P +
Sbjct: 374 --VKLGHVDCDAEKS--LMSRFKVQGFPTI 451
>BG362767
Length = 491
Score = 43.1 bits (100), Expect = 3e-04
Identities = 29/84 (34%), Positives = 41/84 (48%)
Frame = +2
Query: 72 VEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVGHY 131
VEF+A WC C+ P YEKVA FN V ++ VD A K L +K+ V Y
Sbjct: 59 VEFYAPWCGHCKALAPIYEKVAAAFNLDKDV-----VIANVD-ADKYK-DLAEKYGVSGY 217
Query: 132 PMLFWGPPPKFVGGSWEPKQEKSD 155
P L + P G +++ ++ D
Sbjct: 218 PTLKFFPKSNKAGENYDGGRDLDD 289
>TC205812 similar to UP|Q9SEU5 (Q9SEU5) Thioredoxin x, partial (64%)
Length = 904
Score = 40.4 bits (93), Expect = 0.002
Identities = 38/141 (26%), Positives = 57/141 (39%), Gaps = 16/141 (11%)
Frame = +1
Query: 10 LCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYA---------------VELN 54
+CT+ S+S +SS S + + R+ + +T +Y+ E+N
Sbjct: 211 VCTVPASASTSSSVSLTHSWSATARQRLSSHRTRPIFRNYSSAVPKLTVVTCGAAVTEIN 390
Query: 55 ATNF-DAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVD 113
T F D VLK +VEF A WC CR P E +AK + LT V
Sbjct: 391 ETQFKDTVLKANRPV--LVEFVATWCGPCRLISPSMESLAKEYED---------RLTVVK 537
Query: 114 CASKINIKLCDKFSVGHYPML 134
N +L +++ V P L
Sbjct: 538 IDHDANPRLIEEYKVYGLPTL 600
>TC217196 similar to GB|CAD83013.1|45771904|ATH551263 mitochondrial
sulfhydryl oxidase Erv1p {Arabidopsis thaliana;} ,
partial (59%)
Length = 753
Score = 38.9 bits (89), Expect = 0.006
Identities = 21/77 (27%), Positives = 30/77 (38%), Gaps = 3/77 (3%)
Frame = +2
Query: 315 WVLLHSLSVRIEDGESQFTFNAICDFVH---NFFVCEECRQHFYDMCSSVSTPFNKARDF 371
W LH L+ + D + + V + C ECR HF ++ + +F
Sbjct: 215 WTFLHILAAQYPDNPPSHQKKDVKELVQMLPRIYPCGECRDHFKEVLRANPVQTGSHAEF 394
Query: 372 VLWLWSSHNKVNERLSK 388
WL HN VN L K
Sbjct: 395 SQWLCHVHNVVNRSLGK 445
>TC207907 similar to UP|Q96313 (Q96313) AT.I.24-13 protein, partial (80%)
Length = 858
Score = 37.4 bits (85), Expect = 0.016
Identities = 21/86 (24%), Positives = 37/86 (42%)
Frame = +2
Query: 47 HDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGI 106
H + L + F+ +K+ + V+F WC C+N ++ + K G D + G
Sbjct: 149 HSEVITLTSDTFNDKIKEKDTAW-FVKFCVPWCKHCKNLGSLWDDLGKAMEGEDEIEVG- 322
Query: 107 ILLTRVDCASKINIKLCDKFSVGHYP 132
VDC ++ +C K + YP
Sbjct: 323 ----EVDCG--MDKAVCSKVDIHSYP 382
>TC218820 weakly similar to UP|Q8LGB3 (Q8LGB3) Contains similarity to protein
disulfide isomerase-related protein, partial (30%)
Length = 836
Score = 35.8 bits (81), Expect = 0.048
Identities = 23/91 (25%), Positives = 42/91 (45%), Gaps = 2/91 (2%)
Frame = +3
Query: 8 LFLCTISVSSS-ATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDAVLKDTP 66
L LC + S+ A+SSSSS P R + + + ++++ + + D
Sbjct: 96 LLLCMAMIQSTFASSSSSSCLPELPSFRYNLQSQCPISIPSNPPLQVDGNFVEGIFSDIK 275
Query: 67 AT-FAVVEFFANWCPACRNYKPHYEKVAKLF 96
T + + F+A+WCP R P +E ++ F
Sbjct: 276 RTEYTSILFYASWCPFSRKMLPQFEILSSTF 368
>TC217415 similar to UP|O23166 (O23166) Thiol-disulfide interchange like
protein (AT4g37200/C7A10_160) (Thioredoxin-like
protein), partial (66%)
Length = 1100
Score = 35.8 bits (81), Expect = 0.048
Identities = 27/95 (28%), Positives = 42/95 (43%), Gaps = 11/95 (11%)
Frame = +1
Query: 13 ISVSSSATSSSSSSFPGRSILREVHDNDQTG------TTDHDYAVEL-----NATNFDAV 61
+ S+S T+SS P ++I ++ + D+ V L NA ++
Sbjct: 262 VEPSTSTTTSSLPQLPTKNINNKIALVSTLAALGLFLSARLDFGVSLKDLSANAMPYEEA 441
Query: 62 LKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLF 96
L + T VVEF+A+WC CR P KV + F
Sbjct: 442 LSNGKPT--VVEFYADWCEVCRELAPDVYKVEQQF 540
>AW471781
Length = 433
Score = 34.7 bits (78), Expect = 0.11
Identities = 26/97 (26%), Positives = 46/97 (46%), Gaps = 5/97 (5%)
Frame = +1
Query: 53 LNATNFDAVLKDTPATF--AVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLT 110
L+ +NFD+ + A+F +V+F+A WC C+ P + A + A I++
Sbjct: 148 LDESNFDSAI----ASFDHILVDFYAPWCGHCKRLSPELDAAAPVL----ATLKEPIIIA 303
Query: 111 RVDCASKINIKLCDKFSVGHYPMLF---WGPPPKFVG 144
+VD + +L K+ V YP + G P ++ G
Sbjct: 304 KVDADK--HTRLAKKYDVDAYPTILLFNHGVPTEYRG 408
>TC217416 similar to UP|O23166 (O23166) Thiol-disulfide interchange like
protein (AT4g37200/C7A10_160) (Thioredoxin-like
protein), partial (35%)
Length = 827
Score = 34.7 bits (78), Expect = 0.11
Identities = 13/26 (50%), Positives = 17/26 (65%)
Frame = +3
Query: 71 VVEFFANWCPACRNYKPHYEKVAKLF 96
VVEF+A+WC CR + P KV + F
Sbjct: 483 VVEFYADWCEVCREFAPDVYKVEQQF 560
>TC224685 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (55%)
Length = 1169
Score = 34.3 bits (77), Expect = 0.14
Identities = 22/80 (27%), Positives = 40/80 (49%), Gaps = 3/80 (3%)
Frame = +3
Query: 22 SSSSSFPGRSILREVHDNDQTGTTDHDYAVE---LNATNFDAVLKDTPATFAVVEFFANW 78
++ + FP R+ LR D AVE + N+ +++ ++ + +VEF+A W
Sbjct: 390 AAETCFPTRAALRTAPRGGGVKCEAGDTAVEVAPITDANWQSLVLESESP-VLVEFWAPW 566
Query: 79 CPACRNYKPHYEKVAKLFNG 98
C CR P +++AK + G
Sbjct: 567 CGPCRMIHPIIDELAKEYVG 626
>TC224364
Length = 427
Score = 34.3 bits (77), Expect = 0.14
Identities = 24/86 (27%), Positives = 40/86 (45%), Gaps = 2/86 (2%)
Frame = +1
Query: 13 ISVSSSATSSSSSSFP-GRSILREVHDNDQTGTTDHDYAVELNATNFDAVLKDTPAT-FA 70
I+ S SS SS FP S L E+ T+ D ++++ + VL +
Sbjct: 13 ITCLSLLPSSYSSPFPPSASFLYELQSQCSLATSP-DPPLQVDGNFIEGVLSGRKRVGYI 189
Query: 71 VVEFFANWCPACRNYKPHYEKVAKLF 96
+ F+A+WCP R P +E ++ +F
Sbjct: 190SILFYASWCPLSRRMLPEFEILSSMF 267
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,807,325
Number of Sequences: 63676
Number of extensions: 476117
Number of successful extensions: 3185
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 2948
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3116
length of query: 517
length of database: 12,639,632
effective HSP length: 101
effective length of query: 416
effective length of database: 6,208,356
effective search space: 2582676096
effective search space used: 2582676096
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144731.8


