

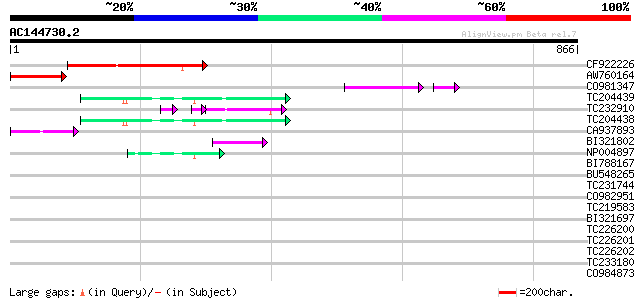
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144730.2 - phase: 0 /pseudo
(866 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922226 188 8e-48
AW760164 similar to GP|11994422|dbj oxidoreductase short-chain ... 93 4e-19
CO981347 83 3e-18
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 67 3e-11
TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial ... 52 7e-11
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 65 1e-10
CA937893 similar to GP|20805072|dbj retrovirus-related pol polyp... 50 3e-06
BI321802 similar to PIR|H72173|H7217 D5L protein - variola minor... 45 1e-04
NP004897 gag-protease polyprotein 43 5e-04
BI788167 42 0.001
BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delal... 40 0.006
TC231744 38 0.006
CO982951 38 0.017
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 38 0.022
BI321697 38 0.022
TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 36 0.064
TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 36 0.064
TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 36 0.064
TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, parti... 35 0.11
CO984873 35 0.11
>CF922226
Length = 667
Score = 188 bits (478), Expect = 8e-48
Identities = 96/218 (44%), Positives = 141/218 (64%), Gaps = 5/218 (2%)
Frame = -3
Query: 89 MTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCAL 148
MTKSL +R KQ LY ++M E + + EQL FNK+I DL NIDV ++DED+AL L C L
Sbjct: 665 MTKSLVNRLYXKQSLYSFKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYL 486
Query: 149 PRSFENFKDTMLYGK*GTITLEEVQAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGK 208
P+S+ +FK+T+L+G+ +++L+EVQ AL +KEL + KE K SGEGL + + +
Sbjct: 485 PKSYSHFKETLLFGR-DSVSLDEVQTALNSKELNERKEKKSSASGEGLTARGKTFKKDSE 309
Query: 209 GKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASN-----E 263
K + +++ G+GN + +C+ C GH +K CPER+ NGG N + + N +
Sbjct: 308 FDKKKQKPENQKNGEGNIFKIRCYHCKKEGHTRKVCPERQKNGGSNNRKKDSGNAAIVQD 129
Query: 264 EGYESAGALTVTSWEPEKGWVLDSGCSYHICPRKEYFE 301
+GYESA AL V+ PE W++DSGCS+H+ P K +FE
Sbjct: 128 DGYESAEALMVSEKNPETKWIMDSGCSWHMTPNKSWFE 15
>AW760164 similar to GP|11994422|dbj oxidoreductase short-chain
dehydrogenase/reductase family-like protein {Arabidopsis
thaliana}, partial (9%)
Length = 428
Score = 93.2 bits (230), Expect = 4e-19
Identities = 47/87 (54%), Positives = 66/87 (75%)
Frame = +3
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAII 60
K+++EKFTG NDFGL +KMRA+L+QQ VEAL GE +++ + +K + KA +AII
Sbjct: 87 KYEVEKFTGQNDFGLC*LKMRALLVQQGLVEALDGEIKLEKMMADGDKKALLQKAYNAII 266
Query: 61 LCLGDKVLREVSRESTAVSMRNKLDSL 87
L LGDKVLR+VS+E+TAV + +KL+ L
Sbjct: 267 LSLGDKVLRQVSKETTAVGVWSKLEVL 347
>CO981347
Length = 624
Score = 82.8 bits (203), Expect(2) = 3e-18
Identities = 56/120 (46%), Positives = 67/120 (55%)
Frame = +3
Query: 512 RRKVMPLKNSKNGVHL*KIRLELN*KC*ELTMVWSLFQSSLMSFAGRKV*RGIELWHTHL 571
+ K KNS+N + L +I L N*K * LTM WSLF SS MSFAG+ +G + TH
Sbjct: 90 KNKSESFKNSENDILLLEINLVQN*KF*GLTMAWSLF*SSSMSFAGK*ASKGTK*SLTHH 269
Query: 572 NRMALQKE*IGLCWSV*GV*CWELDCPKVSGVKLLVLQHI*LTDVHQQG*ISRHLWRFGV 631
+RM QK *I W *G DC + G KL HI*L D Q *+SRH W+ GV
Sbjct: 270 SRMV*QKG*I*PFWKE*GACY*VQDCQRPFGEKLQTQHHI*LIDALHQP*VSRHQWKLGV 449
Score = 28.1 bits (61), Expect(2) = 3e-18
Identities = 18/39 (46%), Positives = 21/39 (53%)
Frame = +1
Query: 648 LLMSGKTNLMLEL*SVFSWVILKV*KVIDCGRWNLEDQN 686
L+M K + M L*SV S ILK K G WNL Q+
Sbjct: 499 LIMLNKESWMQGL*SVCSLAILKELKGTSYGNWNLVRQD 615
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 67.0 bits (162), Expect = 3e-11
Identities = 81/352 (23%), Positives = 135/352 (38%), Gaps = 32/352 (9%)
Frame = +1
Query: 109 VESKPIMEQLTEFNKIIDDL-ANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTI 167
++S+ I++Q + K+I DL A + + E+ + L EN ++ G+
Sbjct: 1135 IKSEKILQQEAQLKKVIADLEAEKEAHKEEISELKGEVGFLNSKLENMTKSIKMLNKGSD 1314
Query: 168 TLEEV---------QAAL----RTKELTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNS 214
TL+EV Q L ++ T E + G +S+ RS++ G + K+
Sbjct: 1315 TLDEVLLLGKNAGNQRGLGFNPKSAGRTTMTEFVPAKNRTGATMSQHRSRHHGMQQKKSK 1494
Query: 215 RSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASNEEGYESAGALTV 274
R K ++C C GH K C G+ G +S+ +
Sbjct: 1495 RKK-----------WRCHYCGKYGHIKPFCYHLHGHP-----------HHGTQSSNSRKK 1608
Query: 275 TSWEPE-----------------KGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNK 317
W P+ + W LDSGCS H+ KE+ +E V G+
Sbjct: 1609 MMWVPKHKAVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGS 1788
Query: 318 ACKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISH- 376
KI +G K+ D L V + L NLISIS G+ + +++
Sbjct: 1789 KGKIIGMG----KLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNE 1956
Query: 377 GALIIAKGSKIHGLYILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGI 428
+ ++ KGS+ L + +S D ++WH R GH+ RG+
Sbjct: 1957 KSEVLMKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGM 2112
>TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial (3%)
Length = 690
Score = 51.6 bits (122), Expect(3) = 7e-11
Identities = 32/130 (24%), Positives = 63/130 (47%), Gaps = 5/130 (3%)
Frame = +2
Query: 299 YFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMF 358
+ E +++G +V +GN I +G + L +F L DV ++P +R+NL+S +
Sbjct: 206 FMEFRPIDDGSIVNMGNVATEPILGLGCVNL-VFTSGKSLYLDVLFVPGIRKNLLSGMIL 382
Query: 359 DGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGST-----VIADASVASVDTLDVT 413
+ G+ +E +S + G + + ++ L + AS +S+ + +
Sbjct: 383 NNCGFKQVLESDKYILSRHGSFVGFGYRCNEMFKLNIDVPFVHESVCMASCSSITNMTKS 562
Query: 414 KLWHLRLGHV 423
++WH RLGHV
Sbjct: 563 EIWHARLGHV 592
Score = 30.4 bits (67), Expect(3) = 7e-11
Identities = 12/26 (46%), Positives = 13/26 (49%)
Frame = +3
Query: 231 CFICHNPGHFKKDCPERKGNGGGNPS 256
C C PGH K+DC KG PS
Sbjct: 33 CSKCGKPGHLKRDCRVFKGKNKAGPS 110
Score = 23.1 bits (48), Expect(3) = 7e-11
Identities = 7/23 (30%), Positives = 12/23 (51%)
Frame = +1
Query: 278 EPEKGWVLDSGCSYHICPRKEYF 300
+ + W DSG + H+C + F
Sbjct: 139 DDDVAWWFDSGATSHVCKDRRLF 207
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 65.1 bits (157), Expect = 1e-10
Identities = 81/352 (23%), Positives = 132/352 (37%), Gaps = 32/352 (9%)
Frame = +1
Query: 109 VESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRS-FENFKDTMLYGK*GTI 167
++S+ I++Q + K+I +L E+E L S EN ++ G+
Sbjct: 1138 IKSEKILQQEAQLKKVIANLEAEKEAHEEEISELKGEVGFLNSKLENMTKSIKMLNKGSD 1317
Query: 168 TLEEV---------QAAL----RTKELTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNS 214
L+EV Q L ++ T E + G +S+ RS++ G + K+
Sbjct: 1318 MLDEVLQLGKNVGNQRGLGFNHKSAGRTTMTEFVPAKNSTGATMSQHRSRHHGTQQKKSK 1497
Query: 215 RSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASNEEGYESAGALTV 274
R K ++C C GH K C G+ G +S+ +
Sbjct: 1498 RKK-----------WRCHYCGKYGHIKPFCYHLHGHP-----------HHGTQSSSSGRK 1611
Query: 275 TSWEPE-----------------KGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNK 317
W P+ + W LDSGCS H+ KE+ +E V G+
Sbjct: 1612 MMWVPKHKIVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGS 1791
Query: 318 ACKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISH- 376
KI +G K+ D L V + L NLISIS G+ + +++
Sbjct: 1792 KGKITGMG----KLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNE 1959
Query: 377 GALIIAKGSKIHGLYILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGI 428
+ ++ KGS+ L + +S D K+WH R GH+ RG+
Sbjct: 1960 KSEVLMKGSRSKDNCYLWTPQETSYSSTCLFSKEDEVKIWHQRFGHLHLRGM 2115
Score = 30.4 bits (67), Expect = 3.5
Identities = 35/124 (28%), Positives = 54/124 (43%)
Frame = +2
Query: 509 TF*RRKVMPLKNSKNGVHL*KIRLELN*KC*ELTMVWSLFQSSLMSFAGRKV*RGIELWH 568
T R PLK S++ V K + ++ + +TM SL +SL++ A K L
Sbjct: 2366 TLSERNQTPLKYSRS*V*DFKEKKTVSSRESGVTMAESLKTASLLNSAHLKASLMSSLQP 2545
Query: 569 THLNRMALQKE*IGLCWSV*GV*CWELDCPKVSGVKLLVLQHI*LTDVHQQG*ISRHLWR 628
H N+MA K GLC + G + P +SG+K T+ H + + H +
Sbjct: 2546 LHHNKMA*LKGKTGLCKKLLGSCFMPKNFPIISGLKP*TQHATSTTESHLEEGLQPHCMK 2725
Query: 629 FGVG 632
G G
Sbjct: 2726 SGKG 2737
>CA937893 similar to GP|20805072|dbj retrovirus-related pol polyprotein from
transposon TNT 1-94-like, partial (7%)
Length = 412
Score = 50.4 bits (119), Expect = 3e-06
Identities = 30/104 (28%), Positives = 56/104 (53%)
Frame = -2
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAII 60
K+++ KF G+ +F LW+ +++ +L Q ++ L+ + E E+ ++ + I
Sbjct: 309 KFEVGKFDGTGNFRLWQKRVKDLLA*QGLLKVLRDSKSNNTEALDWE--EL*ERTATTIR 136
Query: 61 LCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLY 104
LCL D+ L + + + KL+S YM KSL ++ L Q+LY
Sbjct: 135 LCLVDEFLYHMMELAFPGEVWKKLESQYMLKSLTNKLYLMQKLY 4
>BI321802 similar to PIR|H72173|H7217 D5L protein - variola minor virus
(strain Garcia-1966), partial (17%)
Length = 431
Score = 45.4 bits (106), Expect = 1e-04
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 2/86 (2%)
Frame = -3
Query: 311 VCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERG 370
+ +G++K +++ IG +L + LKD +P R+NLIS+S D LGY
Sbjct: 345 IYVGDDKLVEVKAIGHFKLLLCTGFYLDLKDTFVVPSFRQNLISVSYLDKLGYLCSFGNN 166
Query: 371 VMRISHGALIIAKGSKI--HGLYILE 394
+ R+S + I+ S + LY+L+
Sbjct: 165 IFRLSFKSDIVGTRSLLVNDNLYLLD 88
>NP004897 gag-protease polyprotein
Length = 1923
Score = 43.1 bits (100), Expect = 5e-04
Identities = 38/164 (23%), Positives = 61/164 (37%), Gaps = 17/164 (10%)
Frame = +1
Query: 181 LTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHF 240
+T+F K+ G +S+ RS++ G + K+ R K ++C C GH
Sbjct: 1405 MTEFVPAKIST---GATMSQHRSRHHGTQQKKSKRKK-----------WRCHYCGKYGHI 1542
Query: 241 KKDCPERKGNGGGNPSVQIASNEEGYESAGALTVTSWEPE-----------------KGW 283
K C G+ G +S+ + W P+ + W
Sbjct: 1543 KPFCYHLHGHP-----------HHGTQSSSSRRKMMWVPKHKIVSLVVHTSLRASAKEDW 1689
Query: 284 VLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTI 327
LDSGCS H+ KE+ +E V G+ KI +G +
Sbjct: 1690 YLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKL 1821
>BI788167
Length = 421
Score = 42.0 bits (97), Expect = 0.001
Identities = 29/92 (31%), Positives = 48/92 (51%), Gaps = 3/92 (3%)
Frame = +2
Query: 284 VLDSGCS---YHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLK 340
+LD G S +H+ P E F E G V +GN+ A +I +GTI+LKM+D ++
Sbjct: 41 LLDIGFSCATWHMTPC*E*FCTYETILEGFVFMGNDHALEIVGVGTIKLKMYDGIVRTIQ 220
Query: 341 DVRYIPKLRRNLISISMFDGLGYCTRIERGVM 372
V ++ L++N + + D L IE ++
Sbjct: 221 GVLHVKGLKKNQLFVGKLDDLECKIHIEGEIL 316
>BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delalandii},
partial (5%)
Length = 667
Score = 39.7 bits (91), Expect = 0.006
Identities = 23/90 (25%), Positives = 43/90 (47%), Gaps = 3/90 (3%)
Frame = -3
Query: 272 LTVTSWEPEKGWVLDSGCSYHICPRKEYFEML-ELEEGGVVCLGNNKACKIQVIG--TIR 328
LT ++ P + W DSG S+H+ + + + EE + +GN + I G T
Sbjct: 650 LTSSAATPSQSWYPDSGASHHVTNMSQNIQQVAPFEEPIQIIIGNGQGLNINSSGLSTFS 471
Query: 329 LKMFDDRDFLLKDVRYIPKLRRNLISISMF 358
+ +L ++ ++P + +NLI +S F
Sbjct: 470 SPINPQFSLVLSNLLFVPTITKNLIRVSQF 381
>TC231744
Length = 794
Score = 38.1 bits (87), Expect(2) = 0.006
Identities = 45/221 (20%), Positives = 92/221 (41%), Gaps = 3/221 (1%)
Frame = +2
Query: 114 IMEQLTEFNKIIDDLANIDVNLEDEDKALHLPC-ALPRSFENFKDTMLYGK*GTITLEEV 172
I E + E + + L ++ + L ED +HL +LP F FK + K E +
Sbjct: 86 IREYIMEISNLASKLKSLKLEL-GEDLFVHLVLISLPAHFGQFKVSYNTQKDKWSLNELI 262
Query: 173 QAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCF 232
++ +E L + + N R+++++ + K S K + K + ++ C+
Sbjct: 263 SHCVQEEE-----RL*RDRTESAHNKKRKKTKDVAE---KTS*QKKQQKDE----EFTCY 406
Query: 233 ICHNPGHFKKDCPERKGNGGGNPSVQIASNEEGYESAGALTVTSWEPEKGWVLDSGCSYH 292
C H KK CP+ + ++ + ++ P+ W +DSG + H
Sbjct: 407 FCKKSRHMKKKCPKY--------AAWRVKKDKFLTLVCSEVNLAFVPKDTWWVDSGATTH 562
Query: 293 ICPRKE--YFEMLELEEGGVVCLGNNKACKIQVIGTIRLKM 331
I + + L ++ + +G+ K ++ I T RL++
Sbjct: 563 ISMTMQGCLWSRLPSDDERFIFMGDGKKVAVEAIETFRLQL 685
Score = 20.4 bits (41), Expect(2) = 0.006
Identities = 9/15 (60%), Positives = 10/15 (66%)
Frame = +1
Query: 349 RRNLISISMFDGLGY 363
R NLISIS D G+
Sbjct: 736 RGNLISISSLDKFGF 780
>CO982951
Length = 757
Score = 38.1 bits (87), Expect = 0.017
Identities = 19/48 (39%), Positives = 26/48 (53%), Gaps = 1/48 (2%)
Frame = -3
Query: 202 RSQNRGKGKGK-NSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERK 248
+ ++GKG K N+ SK K + C C GHF+KDCP+RK
Sbjct: 308 KKHDKGKGPLKINNNSKQIQKKTSKRNN--CHFCGKSGHFQKDCPKRK 171
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 37.7 bits (86), Expect = 0.022
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Frame = +2
Query: 206 RGKGKGKNSRSKSRSKGDGNKTQY-KCFICHNPGHFKKDCPERKGNGGGN 254
RG+G G+ + R +G G + +C+ C GH +DC +G GGG+
Sbjct: 347 RGRGGGRYGGGEGRGRGFGRRGGGPECYNCGRIGHLARDCYHGQGGGGGD 496
Score = 36.6 bits (83), Expect = 0.049
Identities = 19/57 (33%), Positives = 27/57 (47%)
Frame = +2
Query: 207 GKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASNE 263
G+G G ++R +G G CF C GHF ++CP G G V ++S E
Sbjct: 473 GQGGGGGDDGRNRRRGGGGGGGGGCFNCGEEGHFARECP-NVGKGNE*ECVMVSSIE 640
>BI321697
Length = 368
Score = 37.7 bits (86), Expect = 0.022
Identities = 28/103 (27%), Positives = 44/103 (42%)
Frame = -3
Query: 289 CSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKL 348
C YH C F + + V +GN + G ++L++ +L V +I +
Sbjct: 303 CCYHQCCL*--FPI*QESSTRTVSMGNGSMAHVLGEGQVKLELSSGNFIVLDGVYHISDI 130
Query: 349 RRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLY 391
R+NLIS S+ GY E + I+ I KG GL+
Sbjct: 129 RKNLISTSLLVQQGYKVVFESNRVVITRHGNFIGKGYICDGLF 1
>TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (89%)
Length = 936
Score = 36.2 bits (82), Expect = 0.064
Identities = 17/47 (36%), Positives = 24/47 (50%)
Frame = +2
Query: 205 NRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNG 251
+RG G G+ RS + KC+ C PGHF ++C R G+G
Sbjct: 284 SRGGGGGRGGRSGG--------SDLKCYECGEPGHFARECRMRGGSG 400
>TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 820
Score = 36.2 bits (82), Expect = 0.064
Identities = 17/47 (36%), Positives = 24/47 (50%)
Frame = +1
Query: 205 NRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNG 251
+RG G G+ RS + KC+ C PGHF ++C R G+G
Sbjct: 193 SRGGGGGRGGRSGG--------SDLKCYECGEPGHFARECRMRGGSG 309
>TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 1003
Score = 36.2 bits (82), Expect = 0.064
Identities = 17/47 (36%), Positives = 24/47 (50%)
Frame = +3
Query: 205 NRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNG 251
+RG G G+ RS + KC+ C PGHF ++C R G+G
Sbjct: 780 SRGGGGGRGGRSGG--------SDLKCYECGEPGHFARECRMRGGSG 896
>TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, partial (11%)
Length = 916
Score = 35.4 bits (80), Expect = 0.11
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 1/73 (1%)
Frame = +2
Query: 284 VLDSGCSYHICPRKEYFEMLELEEGGV-VCLGNNKACKIQVIGTIRLKMFDDRDFLLKDV 342
++DSG + H+ P YF G + + N I G I+L+ L +V
Sbjct: 455 IIDSGVTDHMTPHSSYFSSYTFLIGNQHIIVANGSHIPIIGCGNIQLQ----SSLHLNNV 622
Query: 343 RYIPKLRRNLISI 355
Y+PKL NL+SI
Sbjct: 623 LYVPKLSNNLLSI 661
>CO984873
Length = 754
Score = 35.4 bits (80), Expect = 0.11
Identities = 19/54 (35%), Positives = 28/54 (51%), Gaps = 4/54 (7%)
Frame = -2
Query: 199 SRERSQN---RGKGKGKNSRSKSRSKGDGN-KTQYKCFICHNPGHFKKDCPERK 248
+RE+ N G KG +++K + K + KCF C+ GH KKDC + K
Sbjct: 465 NREKRVNLTLHGMKKGDQAKNKGKITAKPVIKNESKCFFCNKKGHIKKDCSKFK 304
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.344 0.151 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,799,109
Number of Sequences: 63676
Number of extensions: 536026
Number of successful extensions: 4878
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 3648
Number of HSP's successfully gapped in prelim test: 142
Number of HSP's that attempted gapping in prelim test: 1092
Number of HSP's gapped (non-prelim): 3929
length of query: 866
length of database: 12,639,632
effective HSP length: 105
effective length of query: 761
effective length of database: 5,953,652
effective search space: 4530729172
effective search space used: 4530729172
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144730.2


