

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
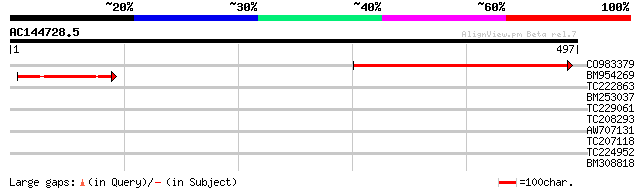
Query= AC144728.5 - phase: 0
(497 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO983379 388 e-108
BM954269 132 4e-31
TC222863 similar to GB|AAP13435.1|30023804|BT006327 At1g20030 {A... 32 0.86
BM253037 31 1.5
TC229061 similar to UP|Q9LR22 (Q9LR22) F26F24.24, partial (37%) 30 3.3
TC208293 similar to UP|O04331 (O04331) Prohibitin 3, partial (44%) 29 4.2
AW707131 homologue to GP|2281647|gb|A AP2 domain containing prot... 28 9.5
TC207118 homologue to UP|Q6T2Z6 (Q6T2Z6) Cyclin d3, complete 28 9.5
TC224952 similar to UP|Q9ZPK0 (Q9ZPK0) Thiosulfate sulfurtransfe... 28 9.5
BM308818 28 9.5
>CO983379
Length = 775
Score = 388 bits (996), Expect = e-108
Identities = 174/192 (90%), Positives = 181/192 (93%)
Frame = -3
Query: 302 DMGSCWKNNGAPCDGDVLTDVTRYSEMIINPETPAWCSPTGLGNCPPFHITPDNKKIFRN 361
D+GSCWKNNGAPCDGDVLTDVTRYSEMIINPETPAWCSP GL NCPPFHITPDNKKI+RN
Sbjct: 773 DIGSCWKNNGAPCDGDVLTDVTRYSEMIINPETPAWCSPKGLVNCPPFHITPDNKKIYRN 594
Query: 362 DTANFPYSAYHYYCAPGNAQHLEQPVSTCDPYSNPQAQEIVQLLPHPIWGEYGYPINKGD 421
DTANFPYSAYHYYCAPGNAQHLEQPVSTCDPYSNP AQEIVQLLPHP WGEYGYP KGD
Sbjct: 593 DTANFPYSAYHYYCAPGNAQHLEQPVSTCDPYSNPHAQEIVQLLPHPTWGEYGYPTKKGD 414
Query: 422 GWVGDARTWELDVGGLASRLYFYQDPGTSPAKRIWTSIDTGTEIFVSDKDEVAEWTISDF 481
GWVGDARTWELDVGGL+SRLYFYQDPGT PAKRIWTSID+GTEIFVSDKDE AEWT+SDF
Sbjct: 413 GWVGDARTWELDVGGLSSRLYFYQDPGTPPAKRIWTSIDSGTEIFVSDKDEEAEWTLSDF 234
Query: 482 DVILIQPKSSLM 493
DVIL Q ++LM
Sbjct: 233 DVILTQSGTNLM 198
>BM954269
Length = 423
Score = 132 bits (332), Expect = 4e-31
Identities = 68/86 (79%), Positives = 75/86 (87%)
Frame = +2
Query: 8 FFVLSNVMLLCASLVSTTQSQTNVSAVGDPGMQRDGLRVAFEAWNFCNEVGQEAPYMGSP 67
F +L NV++ ASLVS+ QS +VSAVGDPGMQRDGLRVAFEAWNFCNEVGQEAP+MGSP
Sbjct: 173 FVLLLNVLVFSASLVSSEQS--HVSAVGDPGMQRDGLRVAFEAWNFCNEVGQEAPHMGSP 346
Query: 68 RAAQCFDLSQGSLIHKVTEKDNKLGV 93
RAA CFDLS GSL HKVTE DN+LGV
Sbjct: 347 RAADCFDLS-GSLTHKVTEADNRLGV 421
>TC222863 similar to GB|AAP13435.1|30023804|BT006327 At1g20030 {Arabidopsis
thaliana;} , partial (75%)
Length = 1057
Score = 31.6 bits (70), Expect = 0.86
Identities = 17/58 (29%), Positives = 22/58 (37%), Gaps = 13/58 (22%)
Frame = -2
Query: 367 PYSAYHYYCAPGNAQHLEQPVSTC-------------DPYSNPQAQEIVQLLPHPIWG 411
PY+ YC NA H + C P+ +P +VQL P P WG
Sbjct: 723 PYAPLQQYCGDPNASHALLHATLCFPSTLVTLSSEGHAPFRSPTHPVLVQLSPEPPWG 550
>BM253037
Length = 237
Score = 30.8 bits (68), Expect = 1.5
Identities = 16/49 (32%), Positives = 20/49 (40%), Gaps = 10/49 (20%)
Frame = -1
Query: 423 WVGDARTWELDVGGLASRLYFYQDPGTSP----------AKRIWTSIDT 461
W+G W L V G S F+Q+ GT R W +IDT
Sbjct: 237 WIGQVHEWRLTVAGQHSDTIFWQEKGTGAKVYIPKHIVFTDRGWQTIDT 91
>TC229061 similar to UP|Q9LR22 (Q9LR22) F26F24.24, partial (37%)
Length = 924
Score = 29.6 bits (65), Expect = 3.3
Identities = 14/43 (32%), Positives = 24/43 (55%)
Frame = -2
Query: 363 TANFPYSAYHYYCAPGNAQHLEQPVSTCDPYSNPQAQEIVQLL 405
TAN ++ H+ +PG+ L PV P+AQ+I+Q++
Sbjct: 215 TANIQHTVSHFRSSPGHCHSLPNPVL-------PKAQQIIQVI 108
>TC208293 similar to UP|O04331 (O04331) Prohibitin 3, partial (44%)
Length = 749
Score = 29.3 bits (64), Expect = 4.2
Identities = 26/78 (33%), Positives = 39/78 (49%), Gaps = 5/78 (6%)
Frame = -3
Query: 233 LFKHKL-KTSKKYPWLMLYLRADTVSGFSG-GYHYDTRGMLKAPLESPNFKVRLTLDVKK 290
+FK+KL KT L D++S S Y Y+ + + A E PNF + L + V+
Sbjct: 705 VFKYKL*KTP-------LSSNKDSISSLSM*FYKYNCKRPIYAQHEGPNFSLILNVKVQI 547
Query: 291 ---GGGSKSQFYLLDMGS 305
+S+FY+L MGS
Sbjct: 546 FH*SSTRRSKFYMLQMGS 493
>AW707131 homologue to GP|2281647|gb|A AP2 domain containing protein RAP2.11
{Arabidopsis thaliana}, partial (22%)
Length = 397
Score = 28.1 bits (61), Expect = 9.5
Identities = 24/101 (23%), Positives = 47/101 (45%), Gaps = 11/101 (10%)
Frame = -2
Query: 213 NNNLSFYEVVWEKKVNVGSWLFKHKLKTSKKYPWLMLYLRADTVS-----------GFSG 261
NNN + + V ++ G W+ + K T K WL Y A+ + G +
Sbjct: 339 NNNTTTSKYVGVRQRASGKWVAEIKDTTQKIRMWLGTYETAEEAARAYDEAACLLRGSNT 160
Query: 262 GYHYDTRGMLKAPLESPNFKVRLTLDVKKGGGSKSQFYLLD 302
++ TR L +PL S +++ ++ +KG +K + Y+++
Sbjct: 159 RTNFITRVSLDSPLAS---RIQNLVNNRKGTKTKQEQYVVE 46
>TC207118 homologue to UP|Q6T2Z6 (Q6T2Z6) Cyclin d3, complete
Length = 1763
Score = 28.1 bits (61), Expect = 9.5
Identities = 16/52 (30%), Positives = 23/52 (43%), Gaps = 9/52 (17%)
Frame = +1
Query: 333 ETPAWCSPTGLGNCPPFHITP---------DNKKIFRNDTANFPYSAYHYYC 375
E +WC P G C P H++P D K IF T +F + ++C
Sbjct: 808 EWSSWCCPPSNGRCTP*HLSPF*ITL*EGLD*KHIF---TGSFSGAVSIFFC 954
>TC224952 similar to UP|Q9ZPK0 (Q9ZPK0) Thiosulfate sulfurtransferase,
partial (95%)
Length = 1758
Score = 28.1 bits (61), Expect = 9.5
Identities = 16/48 (33%), Positives = 26/48 (53%)
Frame = +2
Query: 95 ESLPGLRPEDINNIDLYAVQKELYLGSLCEVKDTPRPWQFWMIMLKNG 142
+ +P R ++ L V LYL ++C+ PRP F+M++ KNG
Sbjct: 14 QRIPK*RNSTLSPHSLVLVIFVLYLVTICDCSSYPRP-LFYMVVNKNG 154
>BM308818
Length = 428
Score = 28.1 bits (61), Expect = 9.5
Identities = 13/24 (54%), Positives = 14/24 (58%), Gaps = 1/24 (4%)
Frame = -2
Query: 331 NPETPAWCSPTGLGN-CPPFHITP 353
NP WC+PT LG P HITP
Sbjct: 265 NPVPKPWCNPTCLGG*RPTSHITP 194
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.138 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,943,754
Number of Sequences: 63676
Number of extensions: 441096
Number of successful extensions: 1868
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1858
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1864
length of query: 497
length of database: 12,639,632
effective HSP length: 101
effective length of query: 396
effective length of database: 6,208,356
effective search space: 2458508976
effective search space used: 2458508976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144728.5


