

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.4 + phase: 0
(566 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
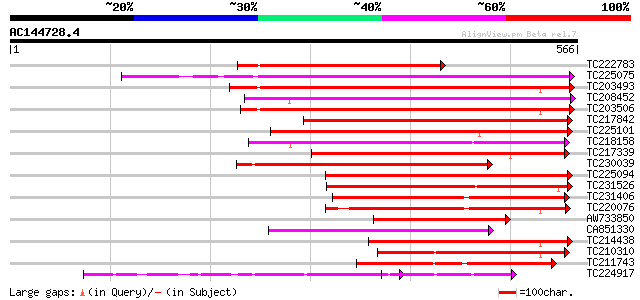
Score E
Sequences producing significant alignments: (bits) Value
TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylester... 338 4e-93
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 332 3e-91
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 311 6e-85
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 298 6e-81
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 297 1e-80
TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 278 4e-75
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 278 5e-75
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 275 5e-74
TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase p... 246 2e-65
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 245 3e-65
TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 231 5e-61
TC231526 similar to UP|PME_MEDSA (Q42920) Pectinesterase precurs... 228 7e-60
TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (... 225 4e-59
TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 pr... 214 8e-56
AW733850 207 1e-53
CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.... 199 4e-51
TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (F... 193 2e-49
TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 192 4e-49
TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 184 7e-47
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 184 9e-47
>TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylesterase
precursor , partial (29%)
Length = 630
Score = 338 bits (867), Expect = 4e-93
Identities = 161/208 (77%), Positives = 187/208 (89%)
Frame = +3
Query: 228 DGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYV 287
+G P W+S ADRKLLA ++ G +V P+A VAKDGSG+F TVLDAINSYPK HQGRY+IYV
Sbjct: 9 EGYPTWVSAADRKLLAQLNDG-AVLPHATVAKDGSGQFTTVLDAINSYPKKHQGRYIIYV 185
Query: 288 KAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAM 347
KAG+YDEYI +DK K N+LIYGDGPTKTIITG+KNF +G KT++TATFSTVAE F+AK++
Sbjct: 186 KAGIYDEYITVDKKKPNLLIYGDGPTKTIITGRKNFHEGTKTMRTATFSTVAEDFMAKSI 365
Query: 348 AFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFG 407
AFENTAGA HQAVALRVQGD+S FFDCA+RGYQDTLYAHAHRQFYRNCEISGT+DFIFG
Sbjct: 366 AFENTAGAEGHQAVALRVQGDRSVFFDCAMRGYQDTLYAHAHRQFYRNCEISGTIDFIFG 545
Query: 408 YASTVIQNSKIVVRKPEANQQNIIVADG 435
Y++T+IQNSKI+VRKP NQQNI+VADG
Sbjct: 546 YSTTLIQNSKILVRKPMPNQQNIVVADG 629
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 332 bits (851), Expect = 3e-91
Identities = 184/457 (40%), Positives = 266/457 (57%), Gaps = 4/457 (0%)
Frame = +1
Query: 112 ALDDCKDLLEFAIDELQASSILAADNSSVHN-VNDRAADLKNWLGAVFAYQQSCLDGFDT 170
A+ DC DLL+ + D L + + + HN + ++DL+ WL A A+ ++C++GF+
Sbjct: 250 AIADCLDLLDLSSDVLSWALSASQNPKGKHNSTGNLSSDLRTWLSAALAHPETCMEGFE- 426
Query: 171 DGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGN 230
G+ V L + + V ++ + L A V P+ + G
Sbjct: 427 ------------GTNSIVKGLVSAGIGQVVSLVEQLLA-----QVLPAQDQFDAASSKGQ 555
Query: 231 -PEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKA 289
P W+ +RKLL ++VTP+ VA DGSG + ++DA+ + P R+VI VK
Sbjct: 556 FPSWIKPKERKLLQ----AIAVTPDVTVALDGSGNYAKIMDAVLAAPDYSMKRFVILVKK 723
Query: 290 GVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAF 349
GVY E ++I K K NI+I G G T+I+G ++ VDG T ++ATF+ GFIA+ ++F
Sbjct: 724 GVYVENVEIKKKKWNIMILGQGMDATVISGNRSVVDGWTTFRSATFAVSGRGFIARDISF 903
Query: 350 ENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYA 409
+NTAG KHQAVALR D S FF C I GYQD+LY H RQF+R+C ISGTVD+IFG A
Sbjct: 904 QNTAGPEKHQAVALRSDSDLSVFFRCGIFGYQDSLYTHTMRQFFRDCTISGTVDYIFGDA 1083
Query: 410 STVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFL 469
+ V QN + V+K NQ+N I A G N PTG Q C I + L P +++L
Sbjct: 1084TAVFQNCFLRVKKGLPNQKNTITAHGRKDPNEPTGFSFQFCNITADSDLIPSVGTAQTYL 1263
Query: 470 ARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-G 528
RPWK+YSR +FM++ + ++I +G+L W G LDT ++AEY NTG G+ V RVKW G
Sbjct: 1264GRPWKSYSRTVFMQSYMSEVIGAEGWLEWNGNFALDTLYYAEYMNTGAGAGVANRVKWPG 1443
Query: 529 KGVLS-KADATKYTAAQWIEGGVWLPATGIPFDLGFT 564
L+ + A+ +T +Q+IEG +WLP+TG+ F G T
Sbjct: 1444YHALNDSSQASNFTVSQFIEGNLWLPSTGVTFTAGLT 1554
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 311 bits (796), Expect = 6e-85
Identities = 154/349 (44%), Positives = 218/349 (62%), Gaps = 4/349 (1%)
Frame = +1
Query: 220 RRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNH 279
RRL + ED P W++G DR+LL+ + + + VV+KDG+G KT+ +AI P+
Sbjct: 109 RRLMAMREDNFPTWLNGRDRRLLSLPLS--QIQADIVVSKDGNGTVKTIAEAIKKVPEYS 282
Query: 280 QGRYVIYVKAGVYDE-YIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTV 338
R +IY++AG Y+E +++ + K N++ GDG KT+ITG +N+ + T TA+F+
Sbjct: 283 SRRIIIYIRAGRYEEDNLKLGRKKTNVMFIGDGKGKTVITGGRNYYQNLTTFHTASFAAS 462
Query: 339 AEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEI 398
GFIAK M FEN AG +HQAVALRV D + + C I GYQDT+Y H++RQFYR C+I
Sbjct: 463 GSGFIAKDMTFENYAGPGRHQAVALRVGADHAVVYRCNIIGYQDTMYVHSNRQFYRECDI 642
Query: 399 SGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPAL 458
GTVDFIFG A+ V QN + RKP A Q+N I A N TG+ + NC IM P L
Sbjct: 643 YGTVDFIFGNAAVVFQNCTLWARKPMAQQKNTITAQNRKDPNQNTGISIHNCRIMATPDL 822
Query: 459 QPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQF-LDTCFFAEYANTGP 517
+ + ++L RPWK Y+R +FM + IGD + P G+L W + F LDTC++ EY N GP
Sbjct: 823 EASKGSYPTYLGRPWKLYARTVFMLSYIGDHVHPRGWLEWNTSSFALDTCYYGEYMNYGP 1002
Query: 518 GSNVQARVKWG--KGVLSKADATKYTAAQWIEGGVWLPATGIPFDLGFT 564
GS + RV W + + S +A+++T Q+I G WLP+TG+ F G +
Sbjct: 1003GSALGQRVNWAGYRAINSTVEASRFTVGQFISGSSWLPSTGVAFIAGLS 1149
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 298 bits (762), Expect = 6e-81
Identities = 153/335 (45%), Positives = 201/335 (59%), Gaps = 4/335 (1%)
Frame = +1
Query: 235 SGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKN---HQGRYVIYVKAGV 291
S +R L S G+ + +V+ G + ++ DAI + P N G +++YV+ G+
Sbjct: 232 SRTERILKESGSQGILLYDFVIVSHYGIDNYTSIGDAIAAAPNNTKPEDGYFLVYVREGL 411
Query: 292 YDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFEN 351
Y+EY+ I K KKNIL+ GDG KTIITG + +DG T ++TF+ E FIA + F N
Sbjct: 412 YEEYVVIPKEKKNILLVGDGINKTIITGNHSVIDGWTTFNSSTFAVSGERFIAVDVTFRN 591
Query: 352 TAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYAST 411
TAG KHQAVA+R D S F+ C+ GYQDTLY H+ RQFYR CEI GTVDFIFG A+
Sbjct: 592 TAGPEKHQAVAVRNNADLSTFYRCSFEGYQDTLYVHSLRQFYRECEIYGTVDFIFGNAAV 771
Query: 412 VIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLAR 471
V Q KI RKP NQ+N + A G N TG+ +QNC I P L D SFL R
Sbjct: 772 VFQGCKIYARKPLPNQKNAVTAQGRTDPNQNTGISIQNCSIDAAPDLVADLNSTMSFLGR 951
Query: 472 PWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKG 530
PWK YSR +++++ IG++IQP G+L W GT LDT F+ E+ N GPGSN RV W G
Sbjct: 952 PWKVYSRTVYLQSYIGNVIQPAGWLEWNGTVGLDTLFYGEFNNYGPGSNTSNRVTWPGYS 1131
Query: 531 VLSKADATKYTAAQWIEGGVWLPATGIPFDLGFTK 565
+L+ A +T + G WLP T IP+ G +
Sbjct: 1132LLNATQAWNFTVLNFTLGNTWLPDTDIPYTEGLLR 1236
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 297 bits (760), Expect = 1e-80
Identities = 147/337 (43%), Positives = 212/337 (62%), Gaps = 3/337 (0%)
Frame = +2
Query: 231 PEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAG 290
P W++ DR+LL+ + ++ + V+K G+G KT+ +AI P++ R++IYV+AG
Sbjct: 14 PGWLNKRDRRLLSLPLS--AIQADITVSKTGNGTVKTIAEAIKKAPEHSTRRFIIYVRAG 187
Query: 291 VYDEY-IQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAF 349
Y+E +++ + K N++ GDG KT+ITGKKN +DG+ T +A+F+ GFIA+ + F
Sbjct: 188 RYEENNLKVGRKKTNVMFIGDGKGKTVITGKKNVIDGMTTFHSASFAASGAGFIARDITF 367
Query: 350 ENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYA 409
EN AG KHQAVALRV D + + C I GYQD+ Y H++RQF+R C I GTVDFIFG A
Sbjct: 368 ENYAGPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDFIFGNA 547
Query: 410 STVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFL 469
+ V Q I RKP A Q+N I A N TG+ L NC I+P P L P + ++L
Sbjct: 548 AVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISLHNCRILPAPDLAPVKGSFPTYL 727
Query: 470 ARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG- 528
RPWK YSR +++ + +GD I P G+L W G L+T ++ EY N GPG+ V RV+W
Sbjct: 728 GRPWKQYSRTVYLVSYMGDHIHPRGWLEWNGDFALNTLYYGEYMNYGPGAAVGQRVQWPG 907
Query: 529 -KGVLSKADATKYTAAQWIEGGVWLPATGIPFDLGFT 564
+ + S +A ++T AQ+I G WLP+TG+ F G +
Sbjct: 908 YRVIKSTMEANRFTVAQFISGSAWLPSTGVAFAAGLS 1018
>TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (48%)
Length = 1225
Score = 278 bits (712), Expect = 4e-75
Identities = 133/270 (49%), Positives = 179/270 (66%), Gaps = 1/270 (0%)
Frame = +3
Query: 294 EYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTA 353
E I I +T KN++I GDG TI+TG N +DG T ++ATF+ +GFIA+ + FENTA
Sbjct: 9 ENIDIKRTVKNLMIVGDGMGATIVTGNHNAIDGSTTFRSATFAVDGDGFIARDITFENTA 188
Query: 354 GANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVI 413
G KHQAVALR D S F+ C+ RGYQDTLY +A+RQFYR+C+I GTVDFIFG A V+
Sbjct: 189 GPQKHQAVALRSGADHSVFYRCSFRGYQDTLYVYANRQFYRDCDIYGTVDFIFGDAVAVL 368
Query: 414 QNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPW 473
QN I VRKP +NQQN + A G N TG+++ NC I L+ + R+FL RPW
Sbjct: 369 QNCNIYVRKPMSNQQNTVTAQGRTDPNENTGIIIHNCRITAAGDLKAVQGSFRTFLGRPW 548
Query: 474 KAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVL 532
+ YSR + M++ + LI P G+ PW+G L T ++AE+ANTG G++ RV W G V+
Sbjct: 549 QKYSRTVVMKSALDGLISPAGWFPWSGNFALSTLYYAEHANTGAGASTGGRVDWAGFRVI 728
Query: 533 SKADATKYTAAQWIEGGVWLPATGIPFDLG 562
S +A K+T ++ GG W+P +G+PFD G
Sbjct: 729 SSTEAVKFTVGNFLAGGSWIPGSGVPFDEG 818
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 278 bits (711), Expect = 5e-75
Identities = 144/307 (46%), Positives = 195/307 (62%), Gaps = 5/307 (1%)
Frame = +3
Query: 261 GSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGK 320
G + V+DA+ + P RYVI++K GVY E ++I K K N+++ GDG TII+G
Sbjct: 207 GRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYENVEIKKKKWNLMMVGDGMDATIISGN 386
Query: 321 KNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGY 380
++F+DG T ++ATF+ GFIA+ + F+NTAG KHQAVALR D S FF C I GY
Sbjct: 387 RSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGY 566
Query: 381 QDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKN 440
QD+LY H RQFYR C+ISGTVDFIFG A+ + QN I +K NQ+N I A G +
Sbjct: 567 QDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHISAKKGLPNQKNTITAHGRKNPD 746
Query: 441 MPTGVVLQNCEIMPEPALQPDRLKVRS---FLARPWKAYSRAIFMENTIGDLIQPDGFLP 497
PTG +Q C I + L S +L RPWK YSR IFM++ I D+++P+G+L
Sbjct: 747 EPTGFSIQFCNISADYDLVNSVNSCNSTHTYLGRPWKPYSRTIFMQSYISDVLRPEGWLE 926
Query: 498 WAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLS-KADATKYTAAQWIEGGVWLPAT 555
W G LDT ++AEY N GPG+ V RVKW G V++ + A+ +T +Q+IEG + LP+T
Sbjct: 927 WNGDFALDTLYYAEYMNYGPGAGVANRVKWQGYHVMNDSSQASNFTVSQFIEGNLCLPST 1106
Query: 556 GIPFDLG 562
G+ F G
Sbjct: 1107GVTFTAG 1127
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 275 bits (702), Expect = 5e-74
Identities = 145/326 (44%), Positives = 192/326 (58%), Gaps = 5/326 (1%)
Frame = +3
Query: 239 RKLL-ADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNH---QGRYVIYVKAGVYDE 294
RKLL A + + V V++DGSG F T+ DAI + P G ++IYV AGVY+E
Sbjct: 102 RKLLQAKVGDEVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSTDGYFLIYVTAGVYEE 281
Query: 295 YIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAG 354
+ IDK K +++ GDG KTIITG ++ VDG T +AT + V +GF+ M NTAG
Sbjct: 282 NVSIDKKKTYLMMVGDGINKTIITGNRSVVDGWTTFSSATLAVVGQGFVGVNMTIRNTAG 461
Query: 355 ANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQ 414
A KHQAVALR D S F+ C+ GYQDTLY H+ RQFY C+I GTVDFIFG A V Q
Sbjct: 462 AVKHQAVALRSGADLSTFYSCSFEGYQDTLYVHSLRQFYSECDIYGTVDFIFGNAKVVFQ 641
Query: 415 NSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWK 474
N K+ R P + Q N I A G N TG+ + NC I L V ++L RPWK
Sbjct: 642 NCKMYPRLPMSGQFNAITAQGRTDPNQDTGISIHNCTIRAADDLAASN-GVATYLGRPWK 818
Query: 475 AYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLS 533
YSR ++M+ + +I G+ W G L T ++AEY+N+GPGS RV W G V++
Sbjct: 819 EYSRTVYMQTVMDSVIHAKGWREWDGDFALSTLYYAEYSNSGPGSGTDNRVTWPGYHVIN 998
Query: 534 KADATKYTAAQWIEGGVWLPATGIPF 559
DA +T + ++ G WLP TG+ +
Sbjct: 999 ATDAANFTVSNFLLGDDWLPQTGVSY 1076
>TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor,
partial (48%)
Length = 923
Score = 246 bits (628), Expect = 2e-65
Identities = 122/262 (46%), Positives = 165/262 (62%), Gaps = 4/262 (1%)
Frame = +1
Query: 302 KKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAV 361
K N+++ GDG T+ITG NF+DG T +TAT + V +GFIA+ + F+NTAG KHQAV
Sbjct: 4 KTNVMLVGDGKDATVITGNLNFIDGTTTFKTATVAAVGDGFIAQDIWFQNTAGPQKHQAV 183
Query: 362 ALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVR 421
ALRV D+S C I +QDTLYAH++RQFYR+ I+GTVDFIFG A+ V Q +V R
Sbjct: 184 ALRVGADQSVINRCRIDAFQDTLYAHSNRQFYRDSFITGTVDFIFGNAAVVFQKCDLVAR 363
Query: 422 KPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIF 481
KP Q N++ A G N TG +Q C + P L+P +++FL RPWK YSR +
Sbjct: 364 KPMDKQNNMVTAQGREDPNQNTGTSIQQCNLTPSSDLKPVVGSIKTFLGRPWKKYSRTVV 543
Query: 482 MENTIGDLIQPDGFLPW--AGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVL-SKADA 537
M++T+ I P G+ W FL T ++ EY N GPG+ RV W G ++ + A+A
Sbjct: 544 MQSTLDSHIDPTGWAEWDAQSKDFLQTLYYGEYMNNGPGAGTSKRVNWPGYHIIKTAAEA 723
Query: 538 TKYTAAQWIEGGVWLPATGIPF 559
+K+T AQ I+G VWL TG+ F
Sbjct: 724 SKFTVAQLIQGNVWLKNTGVNF 789
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 245 bits (626), Expect = 3e-65
Identities = 117/256 (45%), Positives = 167/256 (64%)
Frame = +1
Query: 227 EDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIY 286
+DG P W+S DRKLL + N +VAKDG+G F T+ +A+ P + R+VI+
Sbjct: 151 KDGFPTWLSTKDRKLL--QAAVNETNFNLLVAKDGTGNFTTIAEAVAVAPNSSATRFVIH 324
Query: 287 VKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKA 346
+KAG Y E +++ + K N++ GDG KT++ +N VDG T Q+AT + V +GFIAK
Sbjct: 325 IKAGAYFENVEVIRKKTNLMFVGDGIGKTVVKASRNVVDGWTTFQSATVAVVGDGFIAKG 504
Query: 347 MAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIF 406
+ FEN+AG +KHQAVALR D SAF+ C+ YQDTLY H+ RQFYR+C++ GTVDFIF
Sbjct: 505 ITFENSAGPSKHQAVALRSGSDFSAFYKCSFVAYQDTLYVHSLRQFYRDCDVYGTVDFIF 684
Query: 407 GYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVR 466
G A+TV+QN + RKP NQ+N+ A G N TG+ + NC++ L P + + +
Sbjct: 685 GNAATVLQNCNLYARKPNENQRNLFTAQGREDPNQNTGISILNCKVAAAADLIPVKSQFK 864
Query: 467 SFLARPWKAYSRAIFM 482
++L RPWK YSR +++
Sbjct: 865 NYLGRPWKKYSRTVYL 912
>TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (47%)
Length = 919
Score = 231 bits (590), Expect = 5e-61
Identities = 116/249 (46%), Positives = 162/249 (64%), Gaps = 2/249 (0%)
Frame = +3
Query: 316 IITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDC 375
II+G ++ VDG T ++ATF+ GFIA+ ++F+NTAG KHQAVALR D S FF C
Sbjct: 3 IISGNRSVVDGWTTFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDTDLSVFFRC 182
Query: 376 AIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADG 435
I GYQD+LY H RQF+R C I+GTVD+IFG A+ V QN + V+K NQ+N I A G
Sbjct: 183 GIFGYQDSLYTHTMRQFFRECTITGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHG 362
Query: 436 TVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGF 495
N PTG Q C I + L P +S+L RPWK+YSR +FM++ + ++I+ +G+
Sbjct: 363 RKDPNEPTGFSFQFCNITADSDLVPWVSSTQSYLGRPWKSYSRTVFMQSYMSEVIRGEGW 542
Query: 496 LPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKAD-ATKYTAAQWIEGGVWLP 553
L W G L+T ++ EY NTG G+ + RVKW G + ++ A+ +T AQ+IEG +WLP
Sbjct: 543 LEWNGNFALETLYYGEYMNTGAGAGLANRVKWPGYHPFNDSNQASNFTVAQFIEGNLWLP 722
Query: 554 ATGIPFDLG 562
+TG+ + G
Sbjct: 723 STGVTYTAG 749
>TC231526 similar to UP|PME_MEDSA (Q42920) Pectinesterase precursor (Pectin
methylesterase) (PE) (P65) , partial (56%)
Length = 966
Score = 228 bits (580), Expect = 7e-60
Identities = 110/253 (43%), Positives = 161/253 (63%), Gaps = 7/253 (2%)
Frame = +2
Query: 317 ITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCA 376
ITG KN+VDGV+T TATF A F+AK + FENTAGA KHQAVALRV DK+ F++C
Sbjct: 8 ITGSKNYVDGVQTYNTATFGVNAANFMAKNIGFENTAGAEKHQAVALRVTADKAVFYNCN 187
Query: 377 IRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGT 436
+ G+QDTLY + RQFYR+C ++GT+DF+FG A V QN K +VR P NQQ ++ A G
Sbjct: 188 MDGFQDTLYTQSQRQFYRDCTVTGTIDFVFGDAVAVFQNCKFIVRMPLENQQCLVTAGGR 367
Query: 437 VQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFL 496
+ + P+ +V Q+C EP + K+ ++L RPW+ Y++ + M++ I D+ P+G++
Sbjct: 368 SKIDSPSALVFQSCVFTGEPNVLALTPKI-AYLGRPWRLYAKVVIMDSQIDDIFVPEGYM 544
Query: 497 PWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWI------EGG 549
W G+ F DT + E+ N GPG+N R+ W G VL+ +A +Y ++ E
Sbjct: 545 AWMGSAFKDTSTYYEFNNRGPGANTIGRITWPGFKVLNPIEAVEYYPGKFFQIANSTERD 724
Query: 550 VWLPATGIPFDLG 562
W+ +G+P+ LG
Sbjct: 725 SWILGSGVPYSLG 763
>TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (35%)
Length = 851
Score = 225 bits (574), Expect = 4e-59
Identities = 110/238 (46%), Positives = 148/238 (61%), Gaps = 1/238 (0%)
Frame = +2
Query: 323 FVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQD 382
FVDG T TATF+ FIA+ M F NTAG K QAVAL D++ ++ C I +QD
Sbjct: 2 FVDGTPTFSTATFAVFGRNFIARDMGFRNTAGPQKQQAVALMTSADQAVYYRCQIDAFQD 181
Query: 383 TLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMP 442
+LYAH++RQFYR C I GTVDFIFG ++ V+QN I+ R P QQN I A G NM
Sbjct: 182 SLYAHSNRQFYRECNIYGTVDFIFGNSAVVLQNCNIMPRVPMQGQQNPITAQGKTDPNMN 361
Query: 443 TGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQ 502
TG+ +QNC I P D V+++L RPWK YS +FM + +G I P+G+LPW G
Sbjct: 362 TGISIQNCNITP----FGDLSSVKTYLGRPWKNYSTPVFM*SPMGIFIHPNGWLPWVGNS 529
Query: 503 FLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIPF 559
DT F+AE+ N GPG++ + RV W G V+++ A+ +T ++ G W+ A+G PF
Sbjct: 530 APDTIFYAEFQNVGPGASTKNRVNWKGLRVITRKQASMFTVKAFLSGERWITASGAPF 703
>TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (39%)
Length = 808
Score = 214 bits (545), Expect = 8e-56
Identities = 108/248 (43%), Positives = 152/248 (60%), Gaps = 3/248 (1%)
Frame = +2
Query: 316 IITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDC 375
++TG F VK +GFIAK + F N AGA+KHQAVA R D+S FF C
Sbjct: 50 VLTGSMLFFAAVK----------GKGFIAKDIGFVNNAGASKHQAVAFRSGSDRSVFFRC 199
Query: 376 AIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADG 435
+ G+QDTLYAH++RQFYR+C+I+GT+DFIFG A+ V QN KI+ R+P NQ N I A G
Sbjct: 200 SFNGFQDTLYAHSNRQFYRDCDITGTIDFIFGNAAAVFQNCKIMPRQPLPNQFNTITAQG 379
Query: 436 TVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGF 495
+N TG+++Q + P + L ++L RPWK +S + M++ IG ++P G+
Sbjct: 380 KKDRNQNTGIIIQKSKFTP----LENNLTAPTYLGRPWKDFSTTVIMQSDIGSFLKPVGW 547
Query: 496 LPWA-GTQFLDTCFFAEYANTGPGSNVQARVKWG--KGVLSKADATKYTAAQWIEGGVWL 552
+ W + + T F+AEY NTGPG++V RVKW K L+ +A K+T +I+G WL
Sbjct: 548 MSWVPNVEPVSTIFYAEYQNTGPGADVSQRVKWAGYKPTLTDGEAGKFTVQSFIQGPEWL 727
Query: 553 PATGIPFD 560
P + FD
Sbjct: 728 PNAAVQFD 751
>AW733850
Length = 424
Score = 207 bits (527), Expect = 1e-53
Identities = 92/137 (67%), Positives = 113/137 (82%)
Frame = +1
Query: 364 RVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKP 423
R QGD SA FDCA+ GYQDTLY HA+RQFYRNCEISGT+DFIFG ++T+IQNS+++VRKP
Sbjct: 7 RNQGDMSAMFDCAMHGYQDTLYVHANRQFYRNCEISGTIDFIFGASATLIQNSRVIVRKP 186
Query: 424 EANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFME 483
EANQ N + ADGT QKNM TG+VLQNCEI+PE AL P R + +S+L RPWK ++R + ME
Sbjct: 187 EANQFNTVTADGTKQKNMATGIVLQNCEILPEQALFPSRFQTKSYLGRPWKEFARTVVME 366
Query: 484 NTIGDLIQPDGFLPWAG 500
+ IGD IQP+G+ PW G
Sbjct: 367 SNIGDFIQPEGWTPWDG 417
>CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.150 -
Arabidopsis thaliana, partial (37%)
Length = 700
Score = 199 bits (505), Expect = 4e-51
Identities = 108/226 (47%), Positives = 137/226 (59%), Gaps = 1/226 (0%)
Frame = +2
Query: 259 KDGSGKFKTVLDAINSYPKNHQG-RYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTII 317
K GSG FKTV DA+N+ K + R+VI+VK GVY E I++ NI++ GDG TII
Sbjct: 8 KXGSGNFKTVQDALNAAAKRKEKTRFVIHVKKGVYRENIEVALHNDNIMLVGDGLRNTII 187
Query: 318 TGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAI 377
T ++ DG T +AT FIA+ + F+N+AG +K QAVALR D S F+ C I
Sbjct: 188 TSARSVQDGYTTYSSATAGIDGLHFIARDITFQNSAGVHKGQAVALRSASDLSVFYRCGI 367
Query: 378 RGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTV 437
GYQDTL AHA RQFYR C I GTVDFIFG A+ V QN I R+P Q N+I A G
Sbjct: 368 MGYQDTLMAHAQRQFYRQCYIYGTVDFIFGNAAVVFQNCYIFARRPLEGQANMITAQGRG 547
Query: 438 QKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFME 483
TG+ + N +I P L+P K +FL RPW+ YSR + M+
Sbjct: 548 DPFQNTGISIHNSQIRAAPDLKPVVDKYNTFLGRPWQQYSRVVVMK 685
>TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (Fragment) ,
partial (49%)
Length = 1026
Score = 193 bits (490), Expect = 2e-49
Identities = 95/206 (46%), Positives = 127/206 (61%), Gaps = 2/206 (0%)
Frame = +3
Query: 359 QAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKI 418
QAVALRV GD SAFF+C I +QDTLY H +RQF+ C I+GTVDFIFG ++ V Q+ I
Sbjct: 6 QAVALRVGGDLSAFFNCDILAFQDTLYVHNNRQFFEKCLIAGTVDFIFGNSAVVFQDCDI 185
Query: 419 VVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSR 478
R P + Q+N++ A G V N TG+V+Q C I L+ + +++L RPWK YSR
Sbjct: 186 HARLPSSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATNDLESVKKNFKTYLGRPWKEYSR 365
Query: 479 AIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG--KGVLSKAD 536
+ M+++I D+I P G+ W+G L T + EY NTGPG+ RV W K + A+
Sbjct: 366 TVIMQSSISDVIDPIGWHEWSGNFALSTLVYREYQNTGPGAGTSNRVTWKGYKVITDAAE 545
Query: 537 ATKYTAAQWIEGGVWLPATGIPFDLG 562
A YT +I G WL +TG PF LG
Sbjct: 546 ARDYTPGSFIGGSSWLGSTGFPFSLG 623
>TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (38%)
Length = 678
Score = 192 bits (487), Expect = 4e-49
Identities = 90/194 (46%), Positives = 122/194 (62%), Gaps = 2/194 (1%)
Frame = +1
Query: 368 DKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQ 427
D S F+ C+ GYQDTLY H+ RQFYR C I GTVDFIFG A+ V+QN I R P N+
Sbjct: 1 DLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCNIFARNPP-NK 177
Query: 428 QNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIG 487
N I A G N TG+ + N + L+P + VR++L RPWK YSR +FM+ +
Sbjct: 178 VNTITAQGRTDPNQNTGISIHNSRVTAASDLRPVQNSVRTYLGRPWKQYSRTVFMKTYLD 357
Query: 488 DLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG--KGVLSKADATKYTAAQW 545
LI P G++ W+G LDT ++ EY NTGPGS+ RVKW + + S ++A+K++ A +
Sbjct: 358 GLINPAGWMEWSGNFALDTLYYGEYMNTGPGSSTARRVKWSGYRVITSASEASKFSVANF 537
Query: 546 IEGGVWLPATGIPF 559
I G WLP+T +PF
Sbjct: 538 IAGNAWLPSTKVPF 579
>TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (39%)
Length = 601
Score = 184 bits (468), Expect = 7e-47
Identities = 94/202 (46%), Positives = 125/202 (61%), Gaps = 2/202 (0%)
Frame = +2
Query: 347 MAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIF 406
+ F NTAGA HQAVALR D S F+ C+ GYQDTLY ++ RQFYR C+I GTVDFIF
Sbjct: 8 ITFRNTAGATNHQAVALRSGSDLSVFYRCSFEGYQDTLYVYSDRQFYRECDIYGTVDFIF 187
Query: 407 GYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVR 466
G A+ V QN I R P N+ N I A G N TG+ + N ++ D + VR
Sbjct: 188 GNAAVVFQNCNIYARNP-PNKVNTITAQGRTDPNQNTGISIHNSKV----TAASDLMGVR 352
Query: 467 SFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVK 526
++L RPW+ YSR +FM+ + LI P+G+L W+G L T ++ EY NTGPGS+ RV
Sbjct: 353 TYLGRPWQQYSRTVFMKTYLDSLINPEGWLEWSGNFALSTLYYGEYMNTGPGSSTANRVN 532
Query: 527 W-GKGVLSKA-DATKYTAAQWI 546
W G V++ A +A+K+T +I
Sbjct: 533 WLGYHVITSASEASKFTVGNFI 598
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 184 bits (467), Expect = 9e-47
Identities = 117/322 (36%), Positives = 171/322 (52%), Gaps = 1/322 (0%)
Frame = +2
Query: 74 NDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAIDELQASSIL 133
+D++K ++ ++E ++ T L + N K A DC L E+ I L
Sbjct: 290 SDFLKVSLQLALERAQRSELNTHALGPKCR--NVHEKAAWADCLQLYEYTIQRL------ 445
Query: 134 AADNSSVH-NVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKLT 192
N +++ N D + WL ++C +GF + G D+V L
Sbjct: 446 ---NKTINPNTKCNQTDTQTWLSTALTNLETCKNGF-----------YELGVPDYV--LP 577
Query: 193 ALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMSVT 252
++ +V ++ L+ KP S + +G P W+ DRKLL S+ ++
Sbjct: 578 LMSNNVTKLLSNTLSLNKGPYQYKPPSYK------EGFPTWVKPGDRKLL--QSSSVASN 733
Query: 253 PNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGP 312
N VVAKDGSGK+ TV A+++ PK+ GRYVIYVK+GVY+E Q++ NI++ GDG
Sbjct: 734 ANVVVAKDGSGKYTTVKAAVDAAPKSSSGRYVIYVKSGVYNE--QVEVKGNNIMLVGDGI 907
Query: 313 TKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAF 372
KTIITG K+ G T ++AT + V +GFIA+ + F NTAGA HQAVA R D S F
Sbjct: 908 GKTIITGSKSVGGGTTTFRSATVAAVGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVF 1087
Query: 373 FDCAIRGYQDTLYAHAHRQFYR 394
+ C+ G+QDTLY H+ RQFYR
Sbjct: 1088YRCSFEGFQDTLYVHSERQFYR 1153
Score = 90.1 bits (222), Expect = 2e-18
Identities = 54/137 (39%), Positives = 72/137 (52%), Gaps = 2/137 (1%)
Frame = +3
Query: 372 FFDCAIRGYQDTLYAHAHR-QFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNI 430
F D A+R T Y R C+I GTVDFIFG A+ V+QN I R P Q+ I
Sbjct: 1086 FTDVALR-VSKTHYMFIQRGNSTEECDIYGTVDFIFGNAAAVLQNCNIYARTPP--QRTI 1256
Query: 431 IV-ADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDL 489
V A G N TG+++ N ++ P V+S+L RPW+ YSR +FM+ + L
Sbjct: 1257 TVTAQGRTDPNQNTGIIIHNSKVTGASGFNPS--SVKSYLGRPWQKYSRTVFMKTYLDSL 1430
Query: 490 IQPDGFLPWAGTQFLDT 506
I P G++ W G LDT
Sbjct: 1431 INPAGWMEWDGNFALDT 1481
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,482,598
Number of Sequences: 63676
Number of extensions: 239534
Number of successful extensions: 1134
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 1048
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1054
length of query: 566
length of database: 12,639,632
effective HSP length: 102
effective length of query: 464
effective length of database: 6,144,680
effective search space: 2851131520
effective search space used: 2851131520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144728.4


