

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.10 + phase: 0 /pseudo
(1006 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
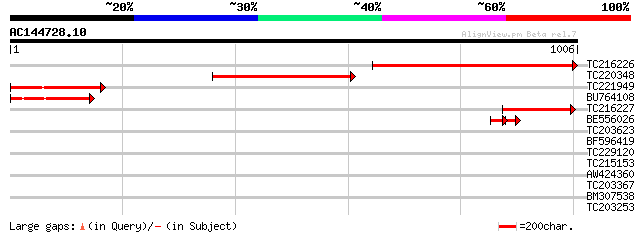
Score E
Sequences producing significant alignments: (bits) Value
TC216226 homologue to UP|SECA_PEA (Q41062) Preprotein translocas... 477 e-134
TC220348 homologue to UP|SECA_PEA (Q41062) Preprotein translocas... 353 3e-97
TC221949 similar to UP|SECA_PEA (Q41062) Preprotein translocase ... 241 9e-64
BU764108 similar to PIR|S65668|S6 preprotein translocase secA pr... 201 2e-51
TC216227 similar to UP|SECA_PEA (Q41062) Preprotein translocase ... 156 5e-38
BE556026 homologue to PIR|S65668|S65 preprotein translocase secA... 46 4e-07
TC203623 similar to UP|Q9SCX3 (Q9SCX3) Elongation factor 1B alph... 33 0.63
BF596419 weakly similar to SP|Q9SYI0|SECA Preprotein translocase... 32 1.1
TC229120 similar to UP|O48726 (O48726) Expressed protein, partia... 32 1.4
TC215153 similar to UP|Q6SZ89 (Q6SZ89) Translational elongation ... 31 2.4
AW424360 similar to SP|P06183|PSBR_ Photosystem II 10 kDa polype... 30 6.9
TC203367 similar to UP|Q6V7X5 (Q6V7X5) 10 kDa photosystem II pol... 30 6.9
BM307538 similar to GP|20196943|gb expressed protein {Arabidopsi... 29 9.0
TC203253 similar to UP|Q6V7X5 (Q6V7X5) 10 kDa photosystem II pol... 29 9.0
>TC216226 homologue to UP|SECA_PEA (Q41062) Preprotein translocase secA
subunit, chloroplast precursor, partial (35%)
Length = 1288
Score = 477 bits (1228), Expect = e-134
Identities = 270/366 (73%), Positives = 291/366 (78%), Gaps = 3/366 (0%)
Frame = +3
Query: 644 ILVKRAQPKMKSLPR*EMHFWKSARNIKSSPRKRGRRSWRLVD*L*WVPSVMNHGELTIS 703
I VKR KMKSLP *EMHFWK +NIKSS RKRGRR W+L D +* SVMNH EL IS
Sbjct: 3 IPVKRVLLKMKSLPS*EMHFWKLEKNIKSSLRKRGRRLWKLADYM**AQSVMNHVELIIS 182
Query: 704 CVVEVADKEIQEAHASF*VLKIIFFGYLAETEFRA**KLSEWKTYLLSLRC*PKH*MKPK 763
CVVEVADKEI+EAHA *VLKI +F +L E EFRA* +LSE KTYLLS RC*PKH* KPK
Sbjct: 183 CVVEVADKEIREAHAFS*VLKITYFVFLVEIEFRA*CELSELKTYLLSPRC*PKH*TKPK 362
Query: 764 RKWRTISLTFGSNCLSMTKYLIAKETEFIQREDEPFNLTIFSLFLLNMLN*Q*MTF*RPT 823
KWRTISLTFGSNCLSM KYL AKE EFIQREDEP NLT FSLFLLNMLN*Q MTF*+ T
Sbjct: 363 GKWRTISLTFGSNCLSMMKYLTAKEIEFIQREDEPLNLTTFSLFLLNMLN*QWMTF*KQT 542
Query: 824 LVLMLQKTAGILIN*LQKSSNIATC*MI*PLIC*GMSVRIMRDCGAISVFAVKKPICKKG 883
LVLMLQKTAGIL N*LQK SNIATC*MI PLIC G V+I R+CG ISVF V KP C+KG
Sbjct: 543 LVLMLQKTAGILKN*LQKFSNIATC*MIYPLICWGTHVQITRNCGIISVFVVVKPTCRKG 722
Query: 884 ILRSNKHRV**KKRSGS*S*AISIGCGKNICKH*NLFSKLWVYEGMRNGIHLLNINLRVT 943
IL SNK + **KK+SGS*S*AISI CGKNIC+H*NLFSK W Y MR+GIHLL+I+L+
Sbjct: 723 ILWSNKQQA**KKQSGS*S*AISIACGKNICRH*NLFSKPWGYGDMRSGIHLLSISLKAI 902
Query: 944 TFSWR*WHK*EEMLYILYISLNPCC*SKIRIKLRIRNQVNG-MHALGTIQ--IQIQLVQL 1000
TFSW *WHK*EEM YILYIS N C *SKIR K RI NQ N +HAL IQ IQ+ L L
Sbjct: 903 TFSWI*WHK*EEMSYILYISSNLCW*SKIRTKQRIGNQENVILHALRLIQTLIQLALSSL 1082
Query: 1001 SHQQVL 1006
HQ +L
Sbjct: 1083 QHQVLL 1100
>TC220348 homologue to UP|SECA_PEA (Q41062) Preprotein translocase secA
subunit, chloroplast precursor, partial (25%)
Length = 776
Score = 353 bits (905), Expect = 3e-97
Identities = 199/253 (78%), Positives = 211/253 (82%)
Frame = +3
Query: 361 FFEM*TT*FAEKRC*LLMSLLVE*CRGEGGVMDFIKQLKQRKGCPSKMKL*HWLLLVTRI 420
FFEM*TT F EKR *LLMSLLVE CRGEGGVM F KQL+QRKGC KMKL* WLL VTRI
Sbjct: 18 FFEM*TTSFVEKRS*LLMSLLVESCRGEGGVMAFTKQLRQRKGCQFKMKL*RWLLSVTRI 197
Query: 421 FFCSSQNYAA*LALHQQKSQNLKAFTSLKLQSCLQTNL**ERMNLM*FSGLLEGNGVQLL 480
F SS N+ *LAL QQK QNL+AFTSLKLQ C QTNL**ERMNLM*F G L+GNGVQL
Sbjct: 198 SFFSSPNFVV*LALQQQKVQNLRAFTSLKLQLCQQTNL**ERMNLM*FLGQLQGNGVQL* 377
Query: 481 WKSLECTKLADLCLLARLALSRVILCLSN*RKLESRTRFLMQNQKMLRGKQKL*HRVVVL 540
KSL CTK CLLA L LSRVILC SN*+KLE RTR+LMQNQKMLRGKQKL*HRVVVL
Sbjct: 378 *KSLGCTKPVARCLLAPLVLSRVILCQSN*KKLEFRTRYLMQNQKMLRGKQKL*HRVVVL 557
Query: 541 GL*QLPPIWLVVGRI*SLAVMLNLWHG*SFVRY*CQELLS*LKEILYR*RNLLLLRHGR* 600
G *QL PIWLVVG+I*SL VMLNLWH *SFVRY*CQELL+ LK+ LY+*RNLL R+GR*
Sbjct: 558 GQ*QLLPIWLVVGQI*SLVVMLNLWHA*SFVRY*CQELLNHLKKALYQ*RNLLPPRYGR* 737
Query: 601 MTSCFHANYQIKI 613
M S FHANYQIK+
Sbjct: 738 MRSYFHANYQIKM 776
>TC221949 similar to UP|SECA_PEA (Q41062) Preprotein translocase secA
subunit, chloroplast precursor, partial (14%)
Length = 655
Score = 241 bits (616), Expect = 9e-64
Identities = 132/172 (76%), Positives = 144/172 (82%), Gaps = 2/172 (1%)
Frame = +2
Query: 1 MAASSLCSSFTSTT--SHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSG 58
MAASSLCSSFTS T H H +H KT + P S+FL + L+ SVS+TRRSR R+SG
Sbjct: 146 MAASSLCSSFTSKTWNPHRPHSCYHHKTPSPPYSTFLRGDVQLHPPSVSRTRRSRRRQSG 325
Query: 59 SVAVASLGGLLGGIFKGNDTGEATRKQYAATVNVINGLEANISKLSDSELRDKTFELRER 118
AVASLGGLLGGIFKG DTGEATR+QYAATVN+INGLE IS LSDSELRD+TF LRER
Sbjct: 326 --AVASLGGLLGGIFKGADTGEATRQQYAATVNIINGLEPEISALSDSELRDRTFALRER 499
Query: 119 AQKRESLDSLLPEAFAVVREASKRVLGLRPFDVQLIGGMVLHKGEIAEMRTG 170
AQ+ +SLDSLLPEAFAVVRE SKRVLGLRPFDVQLIGGMVLHKGEIAEMRTG
Sbjct: 500 AQQGQSLDSLLPEAFAVVREGSKRVLGLRPFDVQLIGGMVLHKGEIAEMRTG 655
>BU764108 similar to PIR|S65668|S6 preprotein translocase secA precursor -
garden pea, partial (14%)
Length = 460
Score = 201 bits (510), Expect = 2e-51
Identities = 113/150 (75%), Positives = 122/150 (81%)
Frame = +3
Query: 1 MAASSLCSSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSGSV 60
MAASSLCSSFTS T + R HH KTL P S+FL + L+ SVS TRR R RRSGSV
Sbjct: 18 MAASSLCSSFTSKTWNPHARYHH-KTLAPPYSAFLRGDLQLHPPSVSGTRRRRRRRSGSV 194
Query: 61 AVASLGGLLGGIFKGNDTGEATRKQYAATVNVINGLEANISKLSDSELRDKTFELRERAQ 120
A SLGGLLGGIFKG DTGEAT++QYAATVN+INGLE IS LSDSELRD+TF LRERAQ
Sbjct: 195 A--SLGGLLGGIFKGADTGEATKQQYAATVNIINGLEPEISALSDSELRDRTFALRERAQ 368
Query: 121 KRESLDSLLPEAFAVVREASKRVLGLRPFD 150
+SLDSLLPEAFAVVREASKRVLGLRPFD
Sbjct: 369 HGQSLDSLLPEAFAVVREASKRVLGLRPFD 458
>TC216227 similar to UP|SECA_PEA (Q41062) Preprotein translocase secA subunit,
chloroplast precursor, partial (13%)
Length = 635
Score = 156 bits (394), Expect = 5e-38
Identities = 88/130 (67%), Positives = 97/130 (73%)
Frame = +3
Query: 875 VKKPICKKGILRSNKHRV**KKRSGS*S*AISIGCGKNICKH*NLFSKLWVYEGMRNGIH 934
V KP C+KGIL SNK + **KK+SGS*S*AISI CGKNIC+H*NLFSK W Y MR+GI
Sbjct: 3 VVKPTCRKGILWSNKQQA**KKQSGS*S*AISIACGKNICRH*NLFSKPWGYGDMRSGIL 182
Query: 935 LLNINLRVTTFSWR*WHK*EEMLYILYISLNPCC*SKIRIKLRIRNQVNGMHALGTIQIQ 994
LL+I+L+ TFSW *WHK*EEM YIL IS N C *SKIR K RI NQ N MH L IQ
Sbjct: 183 LLSISLKAITFSWI*WHK*EEMSYILCISFNLCW*SKIRTKQRIGNQENVMHELRLIQTL 362
Query: 995 IQLVQLSHQQ 1004
IQL S Q+
Sbjct: 363 IQLALSSLQR 392
>BE556026 homologue to PIR|S65668|S65 preprotein translocase secA precursor -
garden pea, partial (2%)
Length = 215
Score = 46.2 bits (108), Expect(2) = 4e-07
Identities = 21/31 (67%), Positives = 23/31 (73%)
Frame = +3
Query: 853 PLIC*GMSVRIMRDCGAISVFAVKKPICKKG 883
PLIC G V+IMR+CG ISVF V KP C KG
Sbjct: 3 PLICWGTRVQIMRNCGIISVFVVVKPTCTKG 95
Score = 26.9 bits (58), Expect(2) = 4e-07
Identities = 17/26 (65%), Positives = 19/26 (72%)
Frame = +2
Query: 880 CKKGILRSNKHRV**KKRSGS*S*AI 905
C IL SNK + **K +SGS*S*AI
Sbjct: 137 CY*RILWSNKQQA**KNQSGS*S*AI 214
>TC203623 similar to UP|Q9SCX3 (Q9SCX3) Elongation factor 1B alpha-subunit,
complete
Length = 899
Score = 33.1 bits (74), Expect = 0.63
Identities = 27/70 (38%), Positives = 37/70 (52%), Gaps = 7/70 (10%)
Frame = -1
Query: 5 SLCSSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLN-------SASVSKTRRSRSRRS 57
++ SSF + S SQ L R+T +LP S FL+ + S+SVS +RSRS S
Sbjct: 536 TVSSSFFMSVSSSQG-LTSRRTEDLPLSFFLAGFLAASLSSAAFLSSSVSSPKRSRSSSS 360
Query: 58 GSVAVASLGG 67
S A+L G
Sbjct: 359 SSAVAAALAG 330
>BF596419 weakly similar to SP|Q9SYI0|SECA Preprotein translocase secA
subunit chloroplast precursor. [Mouse-ear cress]
{Arabidopsis thaliana}, partial (2%)
Length = 406
Score = 32.3 bits (72), Expect = 1.1
Identities = 17/23 (73%), Positives = 19/23 (81%)
Frame = +1
Query: 577 ELLS*LKEILYR*RNLLLLRHGR 599
ELLS LK+ LY+*RNLL RHGR
Sbjct: 4 ELLSHLKKALYQ*RNLLPPRHGR 72
>TC229120 similar to UP|O48726 (O48726) Expressed protein, partial (32%)
Length = 703
Score = 32.0 bits (71), Expect = 1.4
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = -1
Query: 248 ETILPRVSRSLSLGVSIIVSLMRLIQYLSMKLELHSLYQD 287
ETI P+VSR L+ ++ M L+ S+ L + SL+QD
Sbjct: 250 ETIYPQVSRELNAAPKVMCPSMALLDQASIYLRVPSLHQD 131
>TC215153 similar to UP|Q6SZ89 (Q6SZ89) Translational elongation factor 1
subunit Bbeta, complete
Length = 1185
Score = 31.2 bits (69), Expect = 2.4
Identities = 29/84 (34%), Positives = 37/84 (43%), Gaps = 8/84 (9%)
Frame = -3
Query: 8 SSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLN--------SASVSKTRRSRSRRSGS 59
SSF + S S H +T + P S F + F S+SVS +S S S S
Sbjct: 583 SSFFMSVS-SSHGFTSNRTDDFPDSFFFADAFTAAARSSAAFFSSSVSSPNKSTSSSSSS 407
Query: 60 VAVASLGGLLGGIFKGNDTGEATR 83
A A+L GG+ TG ATR
Sbjct: 406 SATAALVSAAGGV----ATGSATR 347
>AW424360 similar to SP|P06183|PSBR_ Photosystem II 10 kDa polypeptide
chloroplast precursor, partial (48%)
Length = 361
Score = 29.6 bits (65), Expect = 6.9
Identities = 16/51 (31%), Positives = 26/51 (50%)
Frame = +1
Query: 132 AFAVVREASKRVLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILPAY 182
+F VV K++ +P+ + GGM L +G A R G+GK + + Y
Sbjct: 214 SFRVVASGGKKIKTDKPYGIN--GGMALREGVDASGRKGKGKGVYQFVDKY 360
>TC203367 similar to UP|Q6V7X5 (Q6V7X5) 10 kDa photosystem II polypeptide,
complete
Length = 759
Score = 29.6 bits (65), Expect = 6.9
Identities = 16/51 (31%), Positives = 26/51 (50%)
Frame = +3
Query: 132 AFAVVREASKRVLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILPAY 182
+F VV K++ +P+ + GGM L +G A R G+GK + + Y
Sbjct: 213 SFRVVASGGKKIKTDKPYGIN--GGMALREGVDASGRKGKGKGVYQFVDKY 359
>BM307538 similar to GP|20196943|gb expressed protein {Arabidopsis thaliana},
partial (35%)
Length = 421
Score = 29.3 bits (64), Expect = 9.0
Identities = 15/40 (37%), Positives = 24/40 (59%)
Frame = -1
Query: 248 ETILPRVSRSLSLGVSIIVSLMRLIQYLSMKLELHSLYQD 287
ETI P+VSR L+ ++ M L+ S+ ++ SL+QD
Sbjct: 268 ETIHPQVSRELNAAPKVMYPSMALLDQASIYPQVPSLHQD 149
>TC203253 similar to UP|Q6V7X5 (Q6V7X5) 10 kDa photosystem II polypeptide,
complete
Length = 1184
Score = 29.3 bits (64), Expect = 9.0
Identities = 16/51 (31%), Positives = 25/51 (48%)
Frame = +2
Query: 132 AFAVVREASKRVLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILPAY 182
+F VV K++ +P+ + GGM L G A R G+GK + + Y
Sbjct: 182 SFRVVASGGKKIKTDKPYGIN--GGMALRDGTDASGRKGKGKGVYQFVDKY 328
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.353 0.155 0.518
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,208,887
Number of Sequences: 63676
Number of extensions: 632747
Number of successful extensions: 7312
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 4085
Number of HSP's successfully gapped in prelim test: 263
Number of HSP's that attempted gapping in prelim test: 3092
Number of HSP's gapped (non-prelim): 4613
length of query: 1006
length of database: 12,639,632
effective HSP length: 107
effective length of query: 899
effective length of database: 5,826,300
effective search space: 5237843700
effective search space used: 5237843700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.5 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144728.10


