

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.1 - phase: 0
(999 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
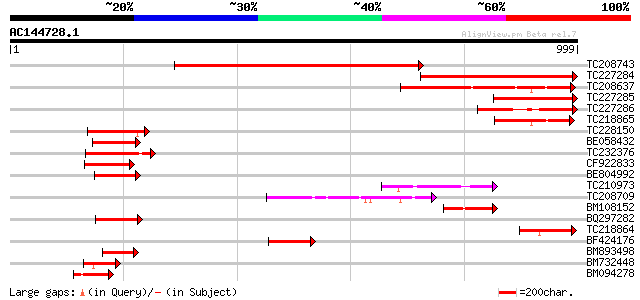
Score E
Sequences producing significant alignments: (bits) Value
TC208743 similar to UP|Q9SMX9 (Q9SMX9) Squamosa promoter binding... 754 0.0
TC227284 similar to UP|O82651 (O82651) Squamosa-promoter binding... 484 e-136
TC208637 similar to UP|O82651 (O82651) Squamosa-promoter binding... 288 8e-78
TC227285 similar to UP|O82651 (O82651) Squamosa-promoter binding... 263 3e-70
TC227286 similar to UP|O82651 (O82651) Squamosa-promoter binding... 231 1e-60
TC218865 similar to UP|O82651 (O82651) Squamosa-promoter binding... 146 4e-35
TC228150 homologue to UP|Q9S849 (Q9S849) Squamosa promoter bindi... 128 1e-29
BE058432 similar to SP|Q38741|SBP1_ Squamosa-promoter binding pr... 128 1e-29
TC232376 125 7e-29
CF922833 121 1e-27
BE804992 similar to PIR|T52297|T522 squamosa promoter binding pr... 112 1e-24
TC210973 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Fr... 111 1e-24
TC208709 similar to UP|Q8S9G8 (Q8S9G8) AT5g18830/F17K4_80, parti... 110 4e-24
BM108152 107 3e-23
BQ297282 similar to PIR|T52607|T526 squamosa promoter binding pr... 105 8e-23
TC218864 similar to UP|O82651 (O82651) Squamosa-promoter binding... 100 3e-21
BF424176 weakly similar to PIR|T52601|T526 squamosa promoter bin... 90 4e-18
BM893498 similar to GP|18650611|gb| At1g27370/F17L21_16 {Arabido... 89 1e-17
BM732448 similar to SP|O04003|LG1_M LIGULELESS1 protein. [Maize]... 81 2e-15
BM094278 74 4e-13
>TC208743 similar to UP|Q9SMX9 (Q9SMX9) Squamosa promoter binding
protein-like 1, partial (35%)
Length = 1326
Score = 754 bits (1947), Expect = 0.0
Identities = 375/441 (85%), Positives = 399/441 (90%), Gaps = 1/441 (0%)
Frame = +3
Query: 290 NLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLIS 349
NLLRE GSSR S M+S LFSNGSQGSPT QH+ VS+ +MQQ+++H HD R +D Q+ S
Sbjct: 3 NLLREDGSSRKSEMMSTLFSNGSQGSPTDTRQHETVSIAKMQQQVMHAHDARAADQQITS 182
Query: 350 SIKPSISNSPPAYSETRDSS-GQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVD 408
SIKPS+SNSPPAYSE RDS+ GQ KMNNFDLNDIY+DSDDG EDLERLPVSTNL TSS+D
Sbjct: 183 SIKPSMSNSPPAYSEARDSTAGQIKMNNFDLNDIYIDSDDGMEDLERLPVSTNLVTSSLD 362
Query: 409 YPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRA 468
YPW QQDSHQSSP QTSGNSDSASAQSPSS SGEAQSRTDRIVFKLFGKEPN+FPLVLRA
Sbjct: 363 YPWAQQDSHQSSPPQTSGNSDSASAQSPSSFSGEAQSRTDRIVFKLFGKEPNDFPLVLRA 542
Query: 469 QILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKT 528
QILDWLS SPTD+ESYIRPGCIVLTIYLRQAEA+WEELC DLTSSL +LLDVSDDTFW+
Sbjct: 543 QILDWLSHSPTDMESYIRPGCIVLTIYLRQAEALWEELCYDLTSSLNRLLDVSDDTFWRN 722
Query: 529 GWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMR 588
GWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKI TVSPIA PASKRAQFSVKGVNL+R
Sbjct: 723 GWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRAQFSVKGVNLIR 902
Query: 589 PATRLMCALEGKYLVCEDAHESTDQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSS 648
PATRLMCALEGKYLVCEDAH S DQ S+E DELQC+QFSCSVPV NGRGFIEIEDQGLSS
Sbjct: 903 PATRLMCALEGKYLVCEDAHMSMDQSSKEPDELQCVQFSCSVPVMNGRGFIEIEDQGLSS 1082
Query: 649 SFFPFIVAEEDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQL 708
SFFPFIV EEDVC+EI LEPLLE SETDPDIEGTGKIKAK+QAMDFIHEMGWLLHRSQL
Sbjct: 1083SFFPFIVVEEDVCSEICTLEPLLELSETDPDIEGTGKIKAKNQAMDFIHEMGWLLHRSQL 1262
Query: 709 KYRMVNLNSGVDLFPLQRFTW 729
K RMV LNS DLFPL+RF W
Sbjct: 1263KLRMVQLNSSEDLFPLKRFKW 1325
>TC227284 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (18%)
Length = 1017
Score = 484 bits (1245), Expect = e-136
Identities = 236/275 (85%), Positives = 251/275 (90%)
Frame = +1
Query: 725 QRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHRAVRRNSKQL 784
+RF WL+EFSMDHDWCA V+KLLNLLLD TVN GDHP+LY ALSEMGLLH+AVRRNSKQL
Sbjct: 1 KRFKWLIEFSMDHDWCAAVRKLLNLLLDGTVNTGDHPSLYLALSEMGLLHKAVRRNSKQL 180
Query: 785 VELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGSEDVLD 844
VE LLRYVP+N SD+LGPEDKALV G+N ++LFRPD G AGLTPLHIAAGKDGSEDVLD
Sbjct: 181 VEWLLRYVPENISDKLGPEDKALVDGENQTFLFRPDVDGTAGLTPLHIAAGKDGSEDVLD 360
Query: 845 ALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAAHVVVEIPS 904
ALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHY YIHLVQKKINK QGAAHVVVEIPS
Sbjct: 361 ALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYAYIHLVQKKINKKQGAAHVVVEIPS 540
Query: 905 NMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRPAMLSMVA 964
NMTE+N N KQNE T EIGK EVRR QG+CKLCD +ISCRTAVGRSMVYRPAMLSMVA
Sbjct: 541 NMTENNTNKKQNELSTIFEIGKPEVRRGQGHCKLCDNRISCRTAVGRSMVYRPAMLSMVA 720
Query: 965 IAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
IAAVCVCVALLFKSSPEV+ MFRPFRWE+LDFGTS
Sbjct: 721 IAAVCVCVALLFKSSPEVICMFRPFRWENLDFGTS 825
>TC208637 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (8%)
Length = 1303
Score = 288 bits (737), Expect = 8e-78
Identities = 158/316 (50%), Positives = 206/316 (65%), Gaps = 7/316 (2%)
Frame = +3
Query: 689 KSQAMDFIHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLN 748
K+QA+DF+ EMGWLLHRS +K+++ ++ DLF RF WL++FSMDH WCAV+KKLL+
Sbjct: 3 KTQALDFLQEMGWLLHRSHVKFKLGSMAPFHDLFQFNRFAWLVDFSMDHGWCAVMKKLLD 182
Query: 749 LLLDETVNKGDHPTLYQALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALV 808
++ + V+ G+H ++ AL MGLLHRAV+RN + +VELLLR+VP TSD E K +
Sbjct: 183 IIFEGGVDAGEHASIELALLNMGLLHRAVKRNCRPMVELLLRFVPVKTSDGADSEMKQVA 362
Query: 809 GGKNHSYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDAL-TNDPCMVGI-EAWKNARDSTG 866
+ +LFRPD VGPAGLTPLH+AA GSE + DP G K ARDSTG
Sbjct: 363 EAPDR-FLFRPDTVGPAGLTPLHVAASMSGSETCVGMH*PYDPSNGGN**HGKCARDSTG 539
Query: 867 STPEDYARLRGHYTYIHLVQKKINKTQGAAHVVVEIPSNMTESNKNPKQNES-----FTS 921
TP D+A LRG+Y+YI LVQ K NK +G +V+IP + N KQ++ S
Sbjct: 540 LTPNDHACLRGYYSYIQLVQNKTNK-KGERQHLVDIPGTVVFFNTTQKQSDGNRTCRVPS 716
Query: 922 LEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPE 981
L+ K E C+ C K++ + +MVYRP MLSMV IA VCVCVALLFKSSP
Sbjct: 717 LKTEKIETTAMPRQCRACQQKVA-YGGMKTAMVYRPVMLSMVTIAVVCVCVALLFKSSPR 893
Query: 982 VLYMFRPFRWESLDFG 997
V Y+F+PF WESL++G
Sbjct: 894 VYYVFQPFNWESLEYG 941
>TC227285 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (10%)
Length = 628
Score = 263 bits (672), Expect = 3e-70
Identities = 129/147 (87%), Positives = 135/147 (91%)
Frame = +1
Query: 853 VGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAAHVVVEIPSNMTESNKN 912
VGIEAWKNARDSTGSTPEDYARLRGHY YIHLVQKKINK QGAAHVVVEIPSN TESN N
Sbjct: 43 VGIEAWKNARDSTGSTPEDYARLRGHYAYIHLVQKKINKRQGAAHVVVEIPSNTTESNTN 222
Query: 913 PKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRPAMLSMVAIAAVCVCV 972
KQNE T+ EIGKAEV R QG+CKLCD +ISCRTAVGRS+VYRPAMLSMVAIAAVCVCV
Sbjct: 223 EKQNELSTTFEIGKAEVIRGQGHCKLCDKRISCRTAVGRSLVYRPAMLSMVAIAAVCVCV 402
Query: 973 ALLFKSSPEVLYMFRPFRWESLDFGTS 999
ALLFKSSPEV+ MFRPFRWE+LDFGTS
Sbjct: 403 ALLFKSSPEVICMFRPFRWENLDFGTS 483
>TC227286 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (13%)
Length = 721
Score = 231 bits (589), Expect = 1e-60
Identities = 123/175 (70%), Positives = 130/175 (74%)
Frame = +3
Query: 825 AGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHL 884
AGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHY YIHL
Sbjct: 3 AGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYAYIHL 182
Query: 885 VQKKINKTQGAAHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKIS 944
VQK KN ++ S + + KLCD +IS
Sbjct: 183 VQK-----------------------KN*QETRSSSC----------GG*DSKLCDNRIS 263
Query: 945 CRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
CRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEV+ MFRPFRWE+LDFGTS
Sbjct: 264 CRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVICMFRPFRWENLDFGTS 428
>TC218865 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (7%)
Length = 457
Score = 146 bits (369), Expect = 4e-35
Identities = 77/146 (52%), Positives = 101/146 (68%), Gaps = 5/146 (3%)
Frame = +3
Query: 854 GIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAAHVVVEIPSNMTESNKNP 913
G EAWK+A+D+TG TP DYA +RG+Y+YI LVQ K + T + HV+ +IP + +SN
Sbjct: 24 GTEAWKSAQDATGLTPYDYASMRGYYSYIQLVQSKTSNTCKSQHVL-DIPGTLVDSNTKQ 200
Query: 914 KQNE-----SFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRPAMLSMVAIAAV 968
KQ++ +SL+ K E C LC K++ + R++VYRPAMLSMVAIAAV
Sbjct: 201 KQSDRHRSSKVSSLQTEKIETTAMPRRCGLCQQKLAYG-GMRRALVYRPAMLSMVAIAAV 377
Query: 969 CVCVALLFKSSPEVLYMFRPFRWESL 994
CVCVALLFKSSP+V Y+F+PF WESL
Sbjct: 378 CVCVALLFKSSPKVYYVFQPFSWESL 455
>TC228150 homologue to UP|Q9S849 (Q9S849) Squamosa promoter binding
protein-like 8, partial (23%)
Length = 685
Score = 128 bits (321), Expect = 1e-29
Identities = 63/121 (52%), Positives = 77/121 (63%), Gaps = 12/121 (9%)
Frame = +1
Query: 137 GAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHI 196
G+ +SN CQ E C ADLS+ K YHRRHKVCE HSKA+ + QRFCQQCSRFH+
Sbjct: 37 GSTISSNSPRCQAEGCNADLSQAKHYHRRHKVCEFHSKAATVIAAGLTQRFCQQCSRFHL 216
Query: 197 LEEFDEGKRSCRRRLAGHNKRRRKTNQ------------EAVPNGSPTNDDQTSSYLLIS 244
L EFD GKRSCR+RLA HN+RRRKT Q E V P + Q+SS + ++
Sbjct: 217 LSEFDNGKRSCRKRLADHNRRRRKTQQPTQEIQKSQSSLENVARSPPDSGAQSSSSVTVA 396
Query: 245 L 245
+
Sbjct: 397 V 399
>BE058432 similar to SP|Q38741|SBP1_ Squamosa-promoter binding protein 1.
[Garden snapdragon] {Antirrhinum majus}, partial (58%)
Length = 433
Score = 128 bits (321), Expect = 1e-29
Identities = 60/84 (71%), Positives = 64/84 (75%)
Frame = +1
Query: 147 CQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRS 206
CQ E CGADL+ K YHRRHKVCE HSKA +V QRFCQQCSRFH L EFDE KRS
Sbjct: 22 CQAERCGADLTDAKRYHRRHKVCEFHSKAPVVVVAGLRQRFCQQCSRFHDLAEFDESKRS 201
Query: 207 CRRRLAGHNKRRRKTNQEAVPNGS 230
CRRRLAGHN+RRRK+N EA GS
Sbjct: 202 CRRRLAGHNERRRKSNPEASNEGS 273
>TC232376
Length = 1033
Score = 125 bits (315), Expect = 7e-29
Identities = 62/124 (50%), Positives = 78/124 (62%)
Frame = +2
Query: 134 KSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSR 193
++R + C V+ C ADLS +DYHRRHKVCE+HSK ++ +G QRFCQQCSR
Sbjct: 2 RARALSNGTQTVSCLVDGCHADLSNCRDYHRRHKVCEVHSKTAQVSIGGQKQRFCQQCSR 181
Query: 194 FHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSNMH 253
FH LEEFDEGKRSCR+RL GHN+RRRK E++ + S+Y LL S+
Sbjct: 182 FHSLEEFDEGKRSCRKRLDGHNRRRRKPQPESLTR----SGSFLSNYQGTQLLPFSSSHE 349
Query: 254 YQPT 257
Y T
Sbjct: 350 YPST 361
>CF922833
Length = 664
Score = 121 bits (304), Expect = 1e-27
Identities = 56/88 (63%), Positives = 63/88 (70%)
Frame = +2
Query: 132 GKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQC 191
GK+S GG S CQV++C ADLS K YHRRHKVCE H+KA + QRFCQQC
Sbjct: 206 GKRSGSKGGGSMPPSCQVDNCDADLSEAKQYHRRHKVCEYHAKAPSVHMAGLQQRFCQQC 385
Query: 192 SRFHILEEFDEGKRSCRRRLAGHNKRRR 219
SRFH L EFD+ KRSCR RLAGHN+RRR
Sbjct: 386 SRFHELSEFDDSKRSCRTRLAGHNERRR 469
>BE804992 similar to PIR|T52297|T522 squamosa promoter binding
protein-homolog 5 [imported] - garden snapdragon
(fragment), partial (22%)
Length = 418
Score = 112 bits (279), Expect = 1e-24
Identities = 50/82 (60%), Positives = 62/82 (74%)
Frame = +2
Query: 149 VEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCR 208
VE C L K+YHRRH+VC+ HSKA +A+V QRFCQQCSRFH++ EFD+ KRSCR
Sbjct: 2 VEGCHVALVNAKEYHRRHRVCDKHSKAPKAVVLGLEQRFCQQCSRFHVVSEFDDSKRSCR 181
Query: 209 RRLAGHNKRRRKTNQEAVPNGS 230
RRLAGHN+RRRK++ +V S
Sbjct: 182 RRLAGHNERRRKSSHLSVTRNS 247
>TC210973 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Fragment),
partial (15%)
Length = 585
Score = 111 bits (278), Expect = 1e-24
Identities = 66/213 (30%), Positives = 116/213 (53%), Gaps = 8/213 (3%)
Frame = +1
Query: 655 VAEEDVCTEIRVLEPLLESSETDPDIEGT------GKIKAKSQAMDFIHEMGWLLHRSQL 708
+A+E +C E++ LE + E D G+ K++ +A+ F++E+GWL R +
Sbjct: 1 IADETICKELKPLESEFDEEEKICDAISEEHEHHFGRPKSREEALHFLNELGWLFQRERF 180
Query: 709 KYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPT-LYQAL 767
Y V + L RF +++ F+++ + C +VK LL++L+ + + T + L
Sbjct: 181 SYV-----HEVPYYSLDRFKFVLTFAVERNCCMLVKTLLDVLVGKHLQGEWLSTGSVEML 345
Query: 768 SEMGLLHRAVRRNSKQLVELLLRY-VPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAG 826
+ + LL+RAV+ +V+LL+ Y +P G + Y+F P+ GP G
Sbjct: 346 NAIQLLNRAVKGKYVGMVDLLIHYSIPSKN-------------GTSRKYVFPPNLEGPGG 486
Query: 827 LTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWK 859
+TPLH+AAG GSE V+D+LT+DP +G++ W+
Sbjct: 487 ITPLHLAAGTSGSESVVDSLTSDPQEIGLKCWE 585
>TC208709 similar to UP|Q8S9G8 (Q8S9G8) AT5g18830/F17K4_80, partial (18%)
Length = 1680
Score = 110 bits (274), Expect = 4e-24
Identities = 87/321 (27%), Positives = 151/321 (46%), Gaps = 22/321 (6%)
Frame = +3
Query: 453 KLFGKEPNEFPLVLRAQILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTS 512
KL+ P EFP LR QI WL+ P ++E YIRPGC +LTI++ +W L D
Sbjct: 3 KLYDWNPAEFPRRLRHQIFQWLASMPVELEGYIRPGCTILTIFIAMPNIMWINLLKDPLE 182
Query: 513 SLIKLLDVSDDTFWK-TGWVHIRVQHQMAF--IFNGQVVIDTSLPFRSNNYSKIWTVSPI 569
+ ++ + T VH+ + M F + +G V + + + K+ V P
Sbjct: 183 YVHDIVAPGKMLSGRGTALVHL---NDMIFRVMKDGTSVTNVKVNMHA---PKLHYVHPT 344
Query: 570 AVPASKRAQFSVKGVNLMRPATRLMCALEGKYLVCEDAHESTDQYSEELDELQCI----Q 625
A K +F G NL++P RL+ + GKYL CE S ++E D + C
Sbjct: 345 YFEAGKPMEFVACGSNLLQPKFRLLVSFSGKYLKCEYCVPSPHSWTE--DNISCAFDNQL 518
Query: 626 FSCSVPVSN----GRGFIEIEDQGLSSSFFPFIVAEEDVCTEIRVLEPLLESSETDPDIE 681
+ VP + G FIE+E++ S+F P ++ ++ +CTE++ L+ L+ S
Sbjct: 519 YKIYVPHTEESLFGPAFIEVENESGLSNFIPVLIGDKKICTEMKTLQQKLDVSLLSKQFR 698
Query: 682 GT--GKI--------KAKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTWLM 731
G I + + + D + ++ WLL + + N + + +QR+ L+
Sbjct: 699 SASGGSICSSCETFALSHTSSSDLLVDIAWLLKDTTSE----NFDRVMTASQIQRYCHLL 866
Query: 732 EFSMDHDWCAVVKKLL-NLLL 751
+F + +D ++ K+L NL++
Sbjct: 867 DFLICNDSTIILGKILPNLII 929
>BM108152
Length = 540
Score = 107 bits (267), Expect = 3e-23
Identities = 53/95 (55%), Positives = 69/95 (71%)
Frame = +1
Query: 765 QALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGP 824
+AL EMGLLH+AV+RN + +VE+LL++VP SD G ++ V ++FRPD VGP
Sbjct: 4 RALLEMGLLHKAVKRNCRPMVEILLKFVPVKASDG-GDSNEKQVNKSPDKFIFRPDTVGP 180
Query: 825 AGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWK 859
GLTPLH+AA GS +VLDALT+DP MVG EAW+
Sbjct: 181 VGLTPLHVAASMHGSXNVLDALTDDPGMVGTEAWE 285
>BQ297282 similar to PIR|T52607|T526 squamosa promoter binding protein 5
[imported] - Arabidopsis thaliana, partial (42%)
Length = 427
Score = 105 bits (263), Expect = 8e-23
Identities = 51/85 (60%), Positives = 57/85 (67%), Gaps = 3/85 (3%)
Frame = +2
Query: 152 CG---ADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCR 208
CG ADL K YHRRH+VCE H KA LV QRFCQQCSRFH L EFD+ KRSCR
Sbjct: 155 CGVVNADLHEAKQYHRRHRVCEYHVKAQVVLVDEVRQRFCQQCSRFHELAEFDDTKRSCR 334
Query: 209 RRLAGHNKRRRKTNQEAVPNGSPTN 233
LAGHN+RRRK + ++ GS N
Sbjct: 335 SSLAGHNERRRKNSDQSQAEGSSRN 409
>TC218864 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (7%)
Length = 821
Score = 100 bits (249), Expect = 3e-21
Identities = 50/104 (48%), Positives = 73/104 (70%), Gaps = 4/104 (3%)
Frame = +3
Query: 899 VVEIPSNMTESNKNPKQNESFTSLEIGKAEVRR----SQGNCKLCDTKISCRTAVGRSMV 954
V++IP N+ +SN KQ++ S ++ + + + +C LC K+ + R++V
Sbjct: 39 VLDIPGNLVDSNTKQKQSDGHRSSKVLSLQTEKIETTAMRHCGLCQQKL-VYGGMRRALV 215
Query: 955 YRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGT 998
+RPAMLSMVAIAAVCVCVALLFKSSP+V Y+F+PF WESL++G+
Sbjct: 216 FRPAMLSMVAIAAVCVCVALLFKSSPKVYYVFQPFSWESLEYGS 347
>BF424176 weakly similar to PIR|T52601|T526 squamosa promoter binding protein
1 [imported] - Arabidopsis thaliana, partial (5%)
Length = 283
Score = 90.1 bits (222), Expect = 4e-18
Identities = 50/87 (57%), Positives = 59/87 (67%), Gaps = 3/87 (3%)
Frame = +2
Query: 456 GKEPNEFPL--VLRAQILDW-LSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTS 512
GKEP F L + + LD +S+ +ESYI PGCIV+TIYLRQAEA+WEELC DLT
Sbjct: 23 GKEPK*FVLSVLYSTRFLDVVISRVLLILESYILPGCIVMTIYLRQAEALWEELCYDLTP 202
Query: 513 SLIKLLDVSDDTFWKTGWVHIRVQHQM 539
S L+DVS DT +TGW I QHQM
Sbjct: 203 SSNMLMDVSHDTISETGWADIMAQHQM 283
>BM893498 similar to GP|18650611|gb| At1g27370/F17L21_16 {Arabidopsis
thaliana}, partial (14%)
Length = 421
Score = 88.6 bits (218), Expect = 1e-17
Identities = 36/63 (57%), Positives = 49/63 (77%)
Frame = +1
Query: 164 RRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQ 223
R+H+VCE HSK+ + ++ +RFCQQCSRFH L EFD+ KRSCR+RL+ HN RRRKT
Sbjct: 1 RKHRVCEAHSKSPKVVIAGLERRFCQQCSRFHALSEFDDKKRSCRQRLSDHNARRRKTQP 180
Query: 224 EAV 226
+++
Sbjct: 181 DSI 189
>BM732448 similar to SP|O04003|LG1_M LIGULELESS1 protein. [Maize] {Zea mays},
partial (12%)
Length = 356
Score = 81.3 bits (199), Expect = 2e-15
Identities = 38/71 (53%), Positives = 45/71 (62%), Gaps = 7/71 (9%)
Frame = +1
Query: 131 DGKKSRGAGGTSNRAV-------CQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNA 183
+ +K +G GG+ R CQ E CGADL+ K YHRRHKVCE+HSKA +V
Sbjct: 139 EDEKKKGGGGSGRRGSSGFFPPSCQAEMCGADLTVAKRYHRRHKVCELHSKAPSVMVAGL 318
Query: 184 MQRFCQQCSRF 194
QRFCQQCSRF
Sbjct: 319 RQRFCQQCSRF 351
>BM094278
Length = 428
Score = 73.6 bits (179), Expect = 4e-13
Identities = 38/71 (53%), Positives = 48/71 (67%), Gaps = 1/71 (1%)
Frame = +2
Query: 113 AGALSLNLAGHVSPVVERDGKKSR-GAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEM 171
A L LNL G + K+ R G+ GTS+ +CQV++C DLS+ KDYHRRHKVCE
Sbjct: 218 APPLQLNLGGRTNN--SNSNKRVRSGSPGTSSYPMCQVDNCREDLSKAKDYHRRHKVCEA 391
Query: 172 HSKASRALVGN 182
HSKAS+AL+ N
Sbjct: 392 HSKASKALLAN 424
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,120,097
Number of Sequences: 63676
Number of extensions: 628783
Number of successful extensions: 3791
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 3697
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3763
length of query: 999
length of database: 12,639,632
effective HSP length: 107
effective length of query: 892
effective length of database: 5,826,300
effective search space: 5197059600
effective search space used: 5197059600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144728.1


