

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.5 + phase: 0
(256 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
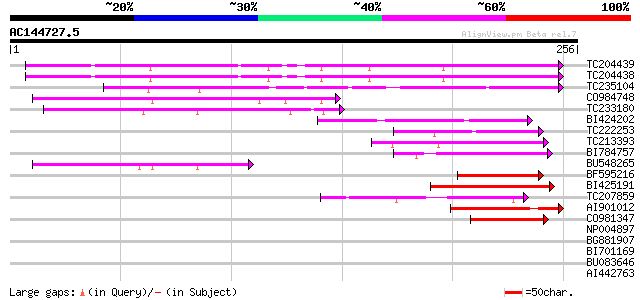
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 111 4e-25
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 110 5e-25
TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotei... 85 3e-17
CO984748 71 4e-13
TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, parti... 68 4e-12
BI424202 63 1e-10
TC222253 similar to UP|Q9ZQE4 (Q9ZQE4) Copia-like retroelement p... 55 2e-08
TC213393 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotei... 55 3e-08
BI784757 51 5e-07
BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delal... 50 1e-06
BF595216 48 5e-06
BI425191 weakly similar to GP|14586969|gb| pol polyprotein {Citr... 47 1e-05
TC207859 46 2e-05
AI901012 44 9e-05
CO981347 42 3e-04
NP004897 gag-protease polyprotein 39 0.002
BG881907 homologue to GP|21432|emb|CA ORF2 {Solanum tuberosum}, ... 39 0.002
BI701169 38 0.004
BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 38 0.004
AI442763 38 0.005
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 111 bits (277), Expect = 4e-25
Identities = 78/256 (30%), Positives = 123/256 (47%), Gaps = 13/256 (5%)
Frame = +1
Query: 8 SNVSRPWFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADGNNLSITDVGDINSD----F 63
++ W+LDSG S HMTG E+L N+ + DG+ I +G + D
Sbjct: 1666 ASAKEDWYLDSGCSRHMTGVKEFLLNIEPC-STSYVTFGDGSKGKIIGMGKLVHDGLPSL 1842
Query: 64 RNVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGR---LFPLQ 120
VL+ GL +NL+S+ QL D NVNF+++ C+V + S +V+ KG + L+ Q
Sbjct: 1843 NKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKS-EVLMKGSRSKDNCYLWTPQ 2019
Query: 121 FISNHLSLPCNNVLNSYED----WHRKLGHPNSTVLSHLFKTGLL-GNKQVVCTASISCL 175
S S C L+S ED WH++ GH + + + G + G + C
Sbjct: 2020 ETS--YSSTC---LSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICG 2184
Query: 176 VCKLAKSKTLPFPSGAHRA-SNCFEMIHSDVWGMSPIASHARYKYFVTFIDDYSRFTWIY 234
C++ K + H+ S E++H D+ G + S +Y +DD+SRFTW+
Sbjct: 2185 ECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVN 2364
Query: 235 FLRSKSEVFSIVITIS 250
F+R KSE F + +S
Sbjct: 2365 FIREKSETFEVFKELS 2412
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 110 bits (276), Expect = 5e-25
Identities = 78/256 (30%), Positives = 123/256 (47%), Gaps = 13/256 (5%)
Frame = +1
Query: 8 SNVSRPWFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADGNNLSITDVGDINSD----F 63
++ W+LDSG S HMTG E+L N+ + DG+ IT +G + D
Sbjct: 1669 ASAKEDWYLDSGCSRHMTGVKEFLVNIEPC-STSYVTFGDGSKGKITGMGKLVHDGLPSL 1845
Query: 64 RNVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGR---LFPLQ 120
VL+ GL +NL+S+ QL D NVNF+++ C+V + S +V+ KG + L+ Q
Sbjct: 1846 NKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKS-EVLMKGSRSKDNCYLWTPQ 2022
Query: 121 FISNHLSLPCNNVLNSYED----WHRKLGHPNSTVLSHLFKTGLL-GNKQVVCTASISCL 175
S S C L S ED WH++ GH + + + G + G + C
Sbjct: 2023 ETS--YSSTC---LFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICG 2187
Query: 176 VCKLAKSKTLPFPSGAHRA-SNCFEMIHSDVWGMSPIASHARYKYFVTFIDDYSRFTWIY 234
C++ K + H+ S E++H D+ G + S +Y +DD+SRFTW+
Sbjct: 2188 ECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVN 2367
Query: 235 FLRSKSEVFSIVITIS 250
F+R KS+ F + +S
Sbjct: 2368 FIREKSDTFEVFKELS 2415
>TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotein
(Fragment), partial (28%)
Length = 865
Score = 85.1 bits (209), Expect = 3e-17
Identities = 61/213 (28%), Positives = 109/213 (50%), Gaps = 5/213 (2%)
Frame = +2
Query: 43 IQIADGNNLSITDVGDINS----DFRNVLVSPGLASNLLSVGQLVD-NNCNVNFSRAGCV 97
I +ADG+ + T +G ++ +V+ G N+ S+ QL NC+V F V
Sbjct: 20 ITLADGSRVVATGIGHVSPTSSLSLNSVVFILGCPFNITSLSQLTRFRNCSVTFDANSFV 199
Query: 98 VQEQVSGKVIAKGPKVGRLFPLQFISNHLSLPCNNVLNSYEDWHRKLGHPNSTVLSHLFK 157
+QE +G I G + L+ ++ +LS C+ V S + H +LGHP+ + L +
Sbjct: 200 IQECGTGWTIGVGIESHGLY---YLKPNLSWVCSAV-TSPKLLHERLGHPHLSKLK-IMV 364
Query: 158 TGLLGNKQVVCTASISCLVCKLAKSKTLPFPSGAHRASNCFEMIHSDVWGMSPIASHARY 217
L K + C + C+L K R + F +IH D+WG + ++S + Y
Sbjct: 365 PSLEKIKDLFCES------CQLGKHVRSSXRHVESRVDSPFLVIHXDIWGPNRVSSMS-Y 523
Query: 218 KYFVTFIDDYSRFTWIYFLRSKSEVFSIVITIS 250
+YFVTFID++S+ T ++ ++ +SE+ S + +++
Sbjct: 524 RYFVTFIDEFSQCTRVFLMKERSEILSFLTSVN 622
>CO984748
Length = 810
Score = 71.2 bits (173), Expect = 4e-13
Identities = 52/177 (29%), Positives = 79/177 (44%), Gaps = 38/177 (21%)
Frame = -2
Query: 11 SRPWFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADGNNLSITDVGDINSDF------- 63
S+ W+++ GA++H+T S + L N GN+Q+ + +G L IT NS F
Sbjct: 575 SQNWYINYGATHHVTASPQNLMNEVPTSGNEQVFLGNGQGLPITGSTVFNSPFASNVKLT 396
Query: 64 -RNVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGP----------- 111
N+L P + NL+SV +N F + CVV+ QV G ++ +G
Sbjct: 395 LNNLLHVPHITKNLVSVS*FAKDNVFFEFHSSHCVVKSQVDGSILLRGALSQDGLYVFSN 216
Query: 112 --------KVGRLFPLQFIS--NHLSLPCNNVLNSYED---------WHRKLGHPNS 149
VG+ +S + LS CNNV S + WH +LGHP+S
Sbjct: 215 LLNSSFKFTVGKAAANVHMSSLSDLSSHCNNVSTSALNTSNYSLVSLWHNRLGHPSS 45
>TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, partial (11%)
Length = 916
Score = 68.2 bits (165), Expect = 4e-12
Identities = 51/154 (33%), Positives = 78/154 (50%), Gaps = 18/154 (11%)
Frame = +2
Query: 16 LDSGASNHMTGSSEYLHNLASYHGNQQIQIADGNNLSITDVGDI----NSDFRNVLVSPG 71
+DSG ++HMT S Y + GNQ I +A+G+++ I G+I + NVL P
Sbjct: 458 IDSGVTDHMTPHSSYFSSYTFLIGNQHIIVANGSHIPIIGCGNIQLQSSLHLNNVLYVPK 637
Query: 72 LASNLLSVGQLV-DNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQFISNH----- 125
L++NLLS+ ++ D NC V F + CV + G++I + G L+ LQ N
Sbjct: 638 LSNNLLSIHKIT*DLNCVVTFFHSHCVF*DLAMGRMIGIAKE*GGLYYLQHEDNKECTR* 817
Query: 126 LSLPCNNVLNSYEDW--------HRKLGHPNSTV 151
+L N+ +S E W H+ LGHP +V
Sbjct: 818 KALTSNHQTSS-EPWSSSQIWLQHKCLGHPPFSV 916
>BI424202
Length = 421
Score = 62.8 bits (151), Expect = 1e-10
Identities = 35/97 (36%), Positives = 50/97 (51%)
Frame = -3
Query: 140 WHRKLGHPNSTVLSHLFKTGLLGNKQVVCTASISCLVCKLAKSKTLPFPSGAHRASNCFE 199
WHR+LGH + + L G+L A V + +T GA R+SN E
Sbjct: 374 WHRRLGHISIERIKRLVNEGVLSTLDF---ADFETYVDCIKGKQTNKSKKGAKRSSNLLE 204
Query: 200 MIHSDVWGMSPIASHARYKYFVTFIDDYSRFTWIYFL 236
+IH+D+ P KYF+TFIDDYSR+ ++YF+
Sbjct: 203 IIHTDI--CCPDMDANSLKYFITFIDDYSRYMYLYFI 99
>TC222253 similar to UP|Q9ZQE4 (Q9ZQE4) Copia-like retroelement pol
polyprotein, partial (4%)
Length = 919
Score = 55.5 bits (132), Expect = 2e-08
Identities = 26/69 (37%), Positives = 41/69 (58%), Gaps = 1/69 (1%)
Frame = +1
Query: 174 CLVCKLAKSKTLPFPSG-AHRASNCFEMIHSDVWGMSPIASHARYKYFVTFIDDYSRFTW 232
C C++ K FP+G + R +++H D+ + I +H YF+TFIDD+S+ W
Sbjct: 331 CDTCEIGKKHRESFPTGKSWRMKKLLKIVHLDLCTVE-IPTHGDNNYFITFIDDFSKKMW 507
Query: 233 IYFLRSKSE 241
+YFL+ KSE
Sbjct: 508 VYFLKQKSE 534
>TC213393 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotein
(Fragment), partial (23%)
Length = 678
Score = 55.1 bits (131), Expect = 3e-08
Identities = 32/85 (37%), Positives = 47/85 (54%), Gaps = 5/85 (5%)
Frame = -2
Query: 164 KQVVCTAS----ISCLVCKLAKSKTLPFPSGAH-RASNCFEMIHSDVWGMSPIASHARYK 218
KQ+V + S +SC C+ K FP R F ++HSDVWG+ +
Sbjct: 389 KQMVSSLSKLPTLSCESCQFGKHVQSSFPYCVICRDRFPFVLVHSDVWGLCHGMPTLESR 210
Query: 219 YFVTFIDDYSRFTWIYFLRSKSEVF 243
YFVTFI+DYS TW++ + ++SE+F
Sbjct: 209 YFVTFIEDYS*QTWLFLMVNRSELF 135
>BI784757
Length = 430
Score = 51.2 bits (121), Expect = 5e-07
Identities = 27/75 (36%), Positives = 42/75 (56%), Gaps = 3/75 (4%)
Frame = +1
Query: 174 CLVCKLAKS---KTLPFPSGAHRASNCFEMIHSDVWGMSPIASHARYKYFVTFIDDYSRF 230
CL CK ++S + +P RA E+I+SDV G S +YF++FID+ +R
Sbjct: 76 CLQCKQSRSTFKQNVPI-----RAKEKLEVIYSDVCGPMQTESLGGNRYFISFIDELTRK 240
Query: 231 TWIYFLRSKSEVFSI 245
W+Y +R KS+ F +
Sbjct: 241 VWVYLIRRKSDFFEV 285
>BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delalandii},
partial (5%)
Length = 667
Score = 49.7 bits (117), Expect = 1e-06
Identities = 35/111 (31%), Positives = 53/111 (47%), Gaps = 11/111 (9%)
Frame = -3
Query: 11 SRPWFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADGNNLSITDVG------DINSDF- 63
S+ W+ DSGAS+H+T S+ + +A + QI I +G L+I G IN F
Sbjct: 626 SQSWYPDSGASHHVTNMSQNIQQVAPFEEPIQIIIGNGQGLNINSSGLSTFSSPINPQFS 447
Query: 64 ---RNVLVSPGLASNLLSVGQLV-DNNCNVNFSRAGCVVQEQVSGKVIAKG 110
N+L P + NL+ V Q DNN F C+ + + V+ +G
Sbjct: 446 LVLSNLLFVPTITKNLIRVSQFCKDNNVYFEFHSYVCLFKS*DTKLVLLEG 294
>BF595216
Length = 421
Score = 47.8 bits (112), Expect = 5e-06
Identities = 19/39 (48%), Positives = 27/39 (68%)
Frame = +2
Query: 203 SDVWGMSPIASHARYKYFVTFIDDYSRFTWIYFLRSKSE 241
SD+ +P S Y Y+VTF+D ++R+TWI+ LR KSE
Sbjct: 8 SDLCSPAPFVSSTGYSYYVTFVDAFTRYTWIFLLRHKSE 124
>BI425191 weakly similar to GP|14586969|gb| pol polyprotein {Citrus x
paradisi}, partial (7%)
Length = 423
Score = 46.6 bits (109), Expect = 1e-05
Identities = 18/56 (32%), Positives = 34/56 (60%)
Frame = +3
Query: 191 AHRASNCFEMIHSDVWGMSPIASHARYKYFVTFIDDYSRFTWIYFLRSKSEVFSIV 246
A R++ E++H+D+ + S + +YF+TFIDDYS + ++Y L K + ++
Sbjct: 81 ATRSTQLLEIMHTDICRPFDVNSFGKERYFITFIDDYSHYGYVYLLHEKFQAVDVL 248
>TC207859
Length = 751
Score = 45.8 bits (107), Expect = 2e-05
Identities = 31/99 (31%), Positives = 47/99 (47%), Gaps = 5/99 (5%)
Frame = +1
Query: 141 HRKLGHPNSTVLSHLFKTGLLGNKQVVCTASIS----CLVCKLAKSKTLPFPSGAHRASN 196
H L HP+ L HL + K + C + + C C +K + P
Sbjct: 481 H*HLHHPSLNKL-HLLVPSVSIIKSLQCESYLLGKHVCHTCSPHVNKHVASP-------- 633
Query: 197 CFEMIHSDVWGMSPIASHARYKYFVTFIDD-YSRFTWIY 234
F ++HSD+WG+S + Y YFVTFIDD + + TW++
Sbjct: 634 -FALVHSDIWGLSCVYLTLGYFYFVTFIDDFFLKCTWVF 747
>AI901012
Length = 410
Score = 43.5 bits (101), Expect = 9e-05
Identities = 22/51 (43%), Positives = 32/51 (62%)
Frame = +1
Query: 200 MIHSDVWGMSPIASHARYKYFVTFIDDYSRFTWIYFLRSKSEVFSIVITIS 250
++HS++WG S I S ++Y+VTFIDD+SR T +F IV+ IS
Sbjct: 79 IVHSNIWGHSCILSILGFRYYVTFIDDFSRHTXTLL---NERMF*IVLCIS 222
>CO981347
Length = 624
Score = 42.0 bits (97), Expect = 3e-04
Identities = 17/35 (48%), Positives = 24/35 (68%)
Frame = +3
Query: 209 SPIASHARYKYFVTFIDDYSRFTWIYFLRSKSEVF 243
S + +H YF+T IDD+SR W+Y L++KSE F
Sbjct: 6 SRVKTHGGSSYFLTIIDDFSRRVWLYVLKNKSESF 110
>NP004897 gag-protease polyprotein
Length = 1923
Score = 39.3 bits (90), Expect = 0.002
Identities = 19/55 (34%), Positives = 28/55 (50%)
Frame = +1
Query: 8 SNVSRPWFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADGNNLSITDVGDINSD 62
++ W+LDSG S HMTG E+L N+ + DG+ IT +G + D
Sbjct: 1669 ASAKEDWYLDSGCSRHMTGVKEFLVNIEPC-STSYVTFGDGSKGKITGMGKLVHD 1830
>BG881907 homologue to GP|21432|emb|CA ORF2 {Solanum tuberosum}, partial (3%)
Length = 429
Score = 39.3 bits (90), Expect = 0.002
Identities = 18/53 (33%), Positives = 27/53 (49%)
Frame = -2
Query: 7 SSNVSRPWFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADGNNLSITDVGDI 59
S + W +DS AS+HMT + N +S H + +QIADG + G +
Sbjct: 179 SKGKRKSWIVDSRASDHMTVDTPVFDNYSSCHDHATVQIADGTLSKVVGKGSV 21
>BI701169
Length = 407
Score = 38.1 bits (87), Expect = 0.004
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 7/42 (16%)
Frame = +2
Query: 220 FVTFIDDYSRFTWIYFLRSKSEVF-------SIVITISGYQV 254
F FIDD+SR TW+YF + K EVF +IV SG+++
Sbjct: 113 FFLFIDDFSRKTWVYFFKHKLEVFENFKKFKAIVEKESGFKI 238
>BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (19%)
Length = 428
Score = 38.1 bits (87), Expect = 0.004
Identities = 32/117 (27%), Positives = 48/117 (40%), Gaps = 7/117 (5%)
Frame = +1
Query: 14 WFLDSGASNHMTGSSEYLHNL-------ASYHGNQQIQIADGNNLSITDVGDINSDFRNV 66
W +DSG +NHMT E L I + ++I + NV
Sbjct: 73 WLIDSGCTNHMTYDRELFTELDEVVFSKVKIRNEAYIDVKGKETVAI*GHTGLKL-ISNV 249
Query: 67 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQFIS 123
L ++ NLLSV QL+ V F C++++ S +V K G F L F++
Sbjct: 250 LYVSEISQNLLSVPQLLKKGYKVLFEDKNCMIKDSESREVFNIQMK-GMSFALDFMN 417
>AI442763
Length = 426
Score = 37.7 bits (86), Expect = 0.005
Identities = 28/93 (30%), Positives = 41/93 (43%), Gaps = 12/93 (12%)
Frame = -2
Query: 163 NKQVVCTASISCLVCKLAKSKTLPFPSGAHRASNC-----------FEMIHSDVWGMSPI 211
NK + S+S + +S L G H C F ++ S++WG S +
Sbjct: 275 NKXPLLVPSVSIIKSPQCESDLL----GKHVCQTCSPHVNKHVGSPFALVPSEIWGFSWV 108
Query: 212 ASHARYKYFVTFIDD-YSRFTWIYFLRSKSEVF 243
Y YFV IDD + + TW+ FL K +VF
Sbjct: 107 YLTLGYFYFVPLIDDFFLKRTWV-FLMKKCDVF 12
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,869,519
Number of Sequences: 63676
Number of extensions: 193248
Number of successful extensions: 1056
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 1042
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1046
length of query: 256
length of database: 12,639,632
effective HSP length: 95
effective length of query: 161
effective length of database: 6,590,412
effective search space: 1061056332
effective search space used: 1061056332
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144727.5


