

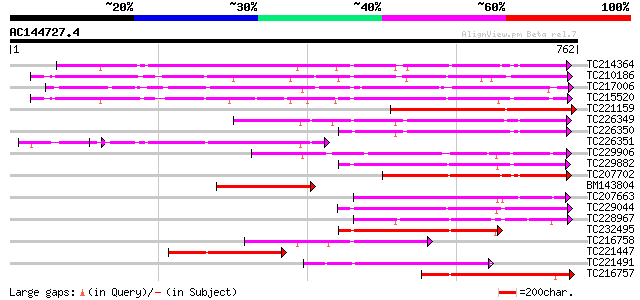
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.4 - phase: 0
(762 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete 455 e-128
TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete 418 e-117
TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete 416 e-116
TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type proteas... 378 e-105
TC221159 weakly similar to UP|Q84TR6 (Q84TR6) Subtilase, partial... 360 1e-99
TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 312 4e-85
TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 228 8e-60
TC226351 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 225 6e-59
TC229906 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like pr... 223 2e-58
TC229882 weakly similar to UP|Q75I27 (Q75I27) Putaive subtilisin... 223 2e-58
TC207702 weakly similar to UP|Q8S896 (Q8S896) Subtilisin-like se... 209 3e-54
BM143804 similar to GP|29028287|gb| subtilase {Casuarina glauca}... 204 2e-52
TC207663 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-... 202 6e-52
TC229044 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease,... 193 2e-49
TC228967 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-li... 193 2e-49
TC232495 similar to UP|Q8W554 (Q8W554) AT3g14240/MLN21_2, partia... 189 5e-48
TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase... 179 4e-45
TC221447 similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like p... 177 1e-44
TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-... 176 3e-44
TC216757 similar to PIR|T05768|T05768 subtilisin-like proteinase... 174 1e-43
>TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete
Length = 2667
Score = 455 bits (1170), Expect = e-128
Identities = 286/726 (39%), Positives = 398/726 (54%), Gaps = 34/726 (4%)
Frame = +1
Query: 64 NNSTSSKIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFL--- 120
+N+ IFY+YT +NGF+A L E + +S + KL TT S F+
Sbjct: 235 SNTAKDSIFYSYTRHINGFAATLDEEVAVEIAKHPKVLSVFENRGRKLHTTRSWDFMELE 414
Query: 121 --GLNPYRGAWPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNI 178
G+ W + FG+ +I+G +DTGVWPES+SF + G+ IPSKW+G +C N
Sbjct: 415 HNGVIQSSSIWKKARFGEGVIIGNLDTGVWPESKSFSEQGLGPIPSKWRG-ICH--NGID 585
Query: 179 QSINLSLCNKKLIGARFFNKGFLAKHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGAS 238
+ + CN+KLIGAR+FNKG+ + ++++ +S RD GHGTHT +TA G+ V S
Sbjct: 586 HTFH---CNRKLIGARYFNKGYASVAGPLNSSF-DSPRDNEGHGTHTLSTAGGNMVARVS 753
Query: 239 FFGYANGTARGIASSSRVAIYKTAWGK--DGDALSSDIIAAIDAAISDGVDILSISLGSD 296
FG +GTA+G + +RVA YK W + +DI+AA D AI DGVD+LS+SLG
Sbjct: 754 VFGQGHGTAKGGSPMARVAAYKVCWPPVAGDECFDADILAAFDLAIHDGVDVLSVSLGGS 933
Query: 297 DLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTV 356
+KD VAI +F A ++G+ V SAGN+GP+ + N PW +TVAA T+DR+F V
Sbjct: 934 SSTFFKDSVAIGSFHAAKRGVVVVCSAGNSGPAEATAENLAPWHVTVAASTMDRQFPTYV 1113
Query: 357 TLGNGVSLTGLSFYLGNFSANNFPIVFMG-------------MCDN-VKELNTVKRKIVV 402
LGN ++ G S + +PI+ +C N + N K KIVV
Sbjct: 1114VLGNDITFKGESLSATKLAHKFYPIIKATDAKLASARAEDAVLCQNGTLDPNKAKGKIVV 1293
Query: 403 CEGNNETLHEQMFNVYKAKVVGGVFISNILDINDV---DNSFPSIIINPVNGEIVKAYIK 459
C ++ + A VG V ++ N++ + P+ IN +G V YI
Sbjct: 1294CLRGINARVDKGEQAFLAGAVGMVLANDKTTGNEIIADPHVLPASHINFTDGSAVFKYI- 1470
Query: 460 SHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAW-- 517
NS +A ++ KT K P + +SS+GP+ P +LKPDITAPG S++AA+
Sbjct: 1471--NSTKFPVAYITHPKTQLDTKPAPFMAAFSSKGPNTIVPEILKPDITAPGVSVIAAYTE 1644
Query: 518 ---PTNVPVSNFGTEVFNN----FNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAI 570
PTN +VF+ FN + GTSMSCPHV+G+ LL+ + WS ++I+SAI
Sbjct: 1645AQGPTN--------QVFDKRRIPFNSVSGTSMSCPHVSGIVGLLRALYPTWSTAAIKSAI 1800
Query: 571 MTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCAL 630
MTT+ LDN E + + +G ATPF+ GAGH+ PNRA+DPGLVYDI + DY+N LCAL
Sbjct: 1801MTTATTLDNEVEPLLNATDGK--ATPFSYGAGHVQPNRAMDPGLVYDITIDDYLNFLCAL 1974
Query: 631 NFTQKNISAITRSSFNDCSKPS-LDLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTT 689
+ + IS T + K S L+LNYPS I S T RT+ NVG T
Sbjct: 1975GYNETQISVFTEGPYKCRKKFSLLNLNYPS-ITVPKLSGSVTVT----RTLKNVG-SPGT 2136
Query: 690 YFASITPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVV 749
Y A + G V+V P+ L FK E+ S+KL + + N AFG L W DGKH V
Sbjct: 2137YIAHVQNPYGITVSVKPSILKFKNVGEEKSFKLTFKAMQGKATNNYAFGKLIWSDGKHYV 2316
Query: 750 RSPIVV 755
SPIVV
Sbjct: 2317TSPIVV 2334
>TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete
Length = 2401
Score = 418 bits (1075), Expect = e-117
Identities = 287/768 (37%), Positives = 409/768 (52%), Gaps = 40/768 (5%)
Frame = +1
Query: 29 YIIHMNLSDMPK-SFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLS 87
YI++M +D SF N H AQV N++ + + Y + +GF+A LS
Sbjct: 181 YIVYMGAADSTDASFRNDH-------AQVL---NSVLRRNENALVRNYKHGFSGFAARLS 330
Query: 88 PEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYR--GAWPTSDFGKDIIVGVIDT 145
+E S+ G +S P LKL TT S FL P + ++G++DT
Sbjct: 331 KKEATSIAQKPGVVSVFPGPVLKLHTTRSWDFLKYQTQVKIDTKPNAVSKSSSVIGILDT 510
Query: 146 GVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHS 205
G+WPE+ SF D GM +PS+WKG + Q S CN+KLIGAR++ +
Sbjct: 511 GIWPEAASFSDKGMGPVPSRWKGTCMKS-----QDFYSSNCNRKLIGARYY--------A 651
Query: 206 NISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGK 265
+ + + N+ RD+NGHGTH + TAAG V AS++G A G A+G + SR+A+Y+
Sbjct: 652 DPNDSGDNTARDSNGHGTHVAGTAAGVMVTNASYYGVATGCAKGGSPESRLAVYRVC--S 825
Query: 266 DGDALSSDIIAAIDAAISDGVDILSISLGSDDLL---LYKDPVAIATFAAMEKGIFVSTS 322
+ S I+AA D AI+DGVD+LS+SLG+ L DP+++ F AME GI V S
Sbjct: 826 NFGCRGSSILAAFDDAIADGVDLLSVSLGASTGFRPDLTSDPISLGAFHAMEHGILVVCS 1005
Query: 323 AGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFS------- 375
AGN+GPS ++ N PW++TVAA T+DR FL + LG+ + G + L S
Sbjct: 1006AGNDGPSSYTLVNDAPWILTVAASTIDRNFLSNIVLGDNKIIKGKAINLSPLSNSPKYPL 1185
Query: 376 -------ANNFPIVFMGMC-DNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVF 427
AN+ +V C N + N VK KIVVC+ N+ + V K VGG+
Sbjct: 1186IYGESAKANSTSLVEARQCHPNSLDGNKVKGKIVVCDDKNDK-YSTRKKVATVKAVGGIG 1362
Query: 428 ISNILDINDVDNS----FPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKST 483
+ +I D N+ S FP+ +I+ +G + YI NS ++ +A + + K
Sbjct: 1363LVHITDQNEAIASNYGDFPATVISSKDGVTILQYI---NSTSNPVATILATTSVLDYKPA 1533
Query: 484 PSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVF------NNFNLI 537
P V +SSRGPS+ +LKPDI APG +ILAAW N GTEV + + +I
Sbjct: 1534PLVPNFSSRGPSSLSSNILKPDIAAPGVNILAAWIGN------GTEVVPKGKKPSLYKII 1695
Query: 538 DGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPF 597
GTSM+CPHV+G+A+ +K + WS SSI+SAIMT++ +N K I ATP+
Sbjct: 1696SGTSMACPHVSGLASSVKTRNPTWSASSIKSAIMTSAIQSNNLKAPITT--ESGSVATPY 1869
Query: 598 ALGAGHINPNRALDPGLVYDIGVQDYINLLCALNF---TQKNISAITRSSFN---DCSKP 651
GAG + + L PGLVY+ DY+N LC + F T K IS +FN D S
Sbjct: 1870DYGAGEMTTSEPLQPGLVYETSSVDYLNFLCYIGFNVTTVKVISKTVPRNFNCPKDLSSD 2049
Query: 652 SL-DLNYPSF-IAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPI-KGFRVTVIPNK 708
+ ++NYPS I FS R RTVTNVGE T ++ I G VT+ PNK
Sbjct: 2050HISNINYPSIAINFSGKR-----AVNLSRTVTNVGEDDETVYSPIVDAPSGVHVTLTPNK 2214
Query: 709 LVFKKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVVT 756
L F K ++K+SY++ I +T + FG ++W +GK++VRSP V+T
Sbjct: 2215LRFTKSSKKLSYRV-IFSSTLTSLKEDLFGSITWSNGKYMVRSPFVLT 2355
>TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete
Length = 2422
Score = 416 bits (1069), Expect = e-116
Identities = 277/735 (37%), Positives = 383/735 (51%), Gaps = 26/735 (3%)
Frame = +3
Query: 49 YESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLP 108
Y S L +V +N + + + + +GF A L+ EE + + ++ P+
Sbjct: 174 YSSMLQEVADSN-----AEPKLVQHHFKRSFSGFVAMLTEEEADRMARHDRVVAVFPNKK 338
Query: 109 LKLDTTHSPQFLGLNPYRGAWPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKG 168
+L TT S F+G P D+I+ V D+G+WPESESF D G PSKWKG
Sbjct: 339 KQLHTTRSWDFIGFPLQANRAPAES---DVIIAVFDSGIWPESESFNDKGFGPPPSKWKG 509
Query: 169 QLCQFENSNIQSINLSLCNKKLIGARFFN-KGFLAKHSNISTTILNSTRDTNGHGTHTST 227
CQ + CN K+IGA+ + GF +K S RD +GHGTH ++
Sbjct: 510 T-CQTSKN-------FTCNNKIIGAKIYKVDGFFSKDDP------KSVRDIDGHGTHVAS 647
Query: 228 TAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVD 287
TAAG+ V AS G GT+RG + +R+A+YK W DG +DI+AA D AI+DGVD
Sbjct: 648 TAAGNPVSTASMLGLGQGTSRGGVTKARIAVYKVCWF-DG-CTDADILAAFDDAIADGVD 821
Query: 288 ILSISLGS-DDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAG 346
I+++SLG D ++D +AI F A+ G+ TSAGN+GP S+ N PW I+VAA
Sbjct: 822 IITVSLGGFSDENYFRDGIAIGAFHAVRNGVLTVTSAGNSGPRPSSLSNFSPWSISVAAS 1001
Query: 347 TLDREFLGTVTLGNGVSLTGLSFYLGNFSANNFPIVFMGMCDNVKE-------------- 392
T+DR+F+ V LGN ++ G S + +PI++ G N E
Sbjct: 1002TIDRKFVTKVELGNKITYEGTSINTFDLKGELYPIIYGGDAPNKGEGIDGSSSRYCSSGS 1181
Query: 393 --LNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDVDNSFPSIIINPVN 450
VK KIV+CE ++ L + A VG + P + +
Sbjct: 1182LDKKLVKGKIVLCESRSKAL-----GPFDAGAVGALIQGQGFRDLPPSLPLPGSYLALQD 1346
Query: 451 GEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPG 510
G V YI NS + IA + FK P V +SSRGP+ P +LKPD+ APG
Sbjct: 1347GASVYDYI---NSTRTPIATI-FKTDETKDTIAPVVASFSSRGPNIVTPEILKPDLVAPG 1514
Query: 511 TSILAAW-PTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSA 569
SILA+W P + P G NFN+I GTSM+CPHV+G AA +K H WSP++IRSA
Sbjct: 1515VSILASWSPASPPSDVEGDNRTLNFNIISGTSMACPHVSGAAAYVKSFHPTWSPAAIRSA 1694
Query: 570 IMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCA 629
+MTT+ L + K H+ FA GAG I+P++A+ PGLVYD G DY+ LC
Sbjct: 1695LMTTAKQL-SPKTHL---------LAEFAYGAGQIDPSKAVYPGLVYDAGEIDYVRFLCG 1844
Query: 630 LNFTQKNISAIT--RSSFNDCSKPSL-DLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEK 686
++ + + IT SS + S DLNY SF F NS+ + F+RTVTNVG
Sbjct: 1845QGYSTRTLQLITGDNSSCPETKNGSARDLNYASFALFVPPYNSNSVSGSFNRTVTNVGSP 2024
Query: 687 KTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKL----KIEGPRMTQKNKVAFGYLSW 742
K+TY A++T KG ++ V P+ L F N+K ++ L K+EGP + G L W
Sbjct: 2025KSTYKATVTSPKGLKIEVNPSVLPFTSLNQKQTFVLTITGKLEGP-------IVSGSLVW 2183
Query: 743 RDGKHVVRSPIVVTN 757
DGK+ VRSPIVV N
Sbjct: 2184DDGKYQVRSPIVVFN 2228
>TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type protease precursor
{Glycine max;} , complete
Length = 2392
Score = 378 bits (970), Expect = e-105
Identities = 276/772 (35%), Positives = 391/772 (49%), Gaps = 44/772 (5%)
Frame = +2
Query: 29 YIIHMNLSDMPK-SFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLS 87
YI++M +D K S N+H AQ+ N++ + + Y + +GF+A LS
Sbjct: 149 YIVYMGAADSTKASLKNEH-------AQIL---NSVLRRNENALVRNYKHGFSGFAARLS 298
Query: 88 PEEHESLKTFSGFISSIPDLPLKLDTTHSPQFL------GLNPYRGAWPTSDFGK-DIIV 140
EE S+ G +S PD LKL TT S FL ++ S F D+I+
Sbjct: 299 KEEANSIAQKPGVVSVFPDPILKLHTTRSWDFLKSQTRVNIDTKPNTLSGSSFSSSDVIL 478
Query: 141 GVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGF 200
GV+DTG+WPE+ SF D G +PS+WKG ++ N S CN+K+IGARF+
Sbjct: 479 GVLDTGIWPEAASFSDKGFGPVPSRWKGTCMTSKD-----FNSSCCNRKIIGARFY---- 631
Query: 201 LAKHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYK 260
+ RD NGHGTH S+TA G V GASF+G A GTARG + SR+A+YK
Sbjct: 632 -------PNPEEKTARDFNGHGTHVSSTAVGVPVSGASFYGLAAGTARGGSPESRLAVYK 790
Query: 261 TAWGKDGDALSSDIIAAIDAAISDGVDILSISL---GSDDLLLYKDPVAIATFAAMEKGI 317
G G S I+A D AI DGVDILS+SL G L DP+AI F ++++GI
Sbjct: 791 VC-GAFGSCPGSAILAGFDDAIHDGVDILSLSLGGFGGTKTDLTTDPIAIGAFHSVQRGI 967
Query: 318 FVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFS-- 375
V +AGN+G F ++ N PW++TVAA T+DR+ V LGN + G + NFS
Sbjct: 968 LVVCAAGNDGEPF-TVLNDAPWILTVAASTIDRDLQSDVVLGNNQVVKGRAI---NFSPL 1135
Query: 376 --ANNFPIVFMGMC--DNVKELNTVKR-------------KIVVCEGNNETLHEQMFNVY 418
+ ++P+++ N+ + ++ KIVVC+G N+ + +
Sbjct: 1136LNSPDYPMIYAESAARANISNITDARQCHPDSLDPKKVIGKIVVCDGKNDIYYSTDEKIV 1315
Query: 419 KAKVVGGVFISNILDIND------VDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMS 472
K +GG+ + +I D + VD FP + +G+ + YI NS + + +
Sbjct: 1316IVKALGGIGLVHITDQSGSVAFYYVD--FPVTEVKSKHGDAILQYI---NSTSHPVGTIL 1480
Query: 473 FKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFN 532
T K P V ++SSRGPS VLKPDI APG +ILAAW N +
Sbjct: 1481ATVTIPDYKPAPRVGYFSSRGPSLITSNVLKPDIAAPGVNILAAWFGNDTSEVPKGRKPS 1660
Query: 533 NFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNR 592
+ ++ GTSM+ PHV+G+A +K + WS S+I+SAIMT++ DN K I +
Sbjct: 1661LYRILSGTSMATPHVSGLACSVKRKNPTWSASAIKSAIMTSAIQNDNLKGPITT--DSGL 1834
Query: 593 AATPFALGAGHINPNRALDPGLVYDIGVQDYINLLC--ALNFTQ-KNISAITRSSFNDCS 649
ATP+ GAG I + L PGLVY+ DY+N LC LN T K IS +FN
Sbjct: 1835IATPYDYGAGAITTSEPLQPGLVYETNNVDYLNYLCYNGLNITMIKVISGTVPENFNCPK 2014
Query: 650 KPSLDL----NYPSFIAFSNARNSSRTTNEFHRTVTNVGEK-KTTYFASITPIKGFRVTV 704
S DL NYPS + + + RTVTNV E+ +T YF + VT+
Sbjct: 2015DSSSDLISSINYPSIAVNFTGKADAVVS----RTVTNVDEEDETVYFPVVEAPSEVIVTL 2182
Query: 705 IPNKLVFKKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVVT 756
P L F +K SY + P+ T K FG ++W + K++VR P V+T
Sbjct: 2183FPYNLEFTTSIKKQSYNITFR-PK-TSLKKDLFGSITWSNDKYMVRIPFVLT 2332
>TC221159 weakly similar to UP|Q84TR6 (Q84TR6) Subtilase, partial (16%)
Length = 855
Score = 360 bits (924), Expect = 1e-99
Identities = 175/251 (69%), Positives = 210/251 (82%), Gaps = 1/251 (0%)
Frame = +3
Query: 512 SILAAWPTNVPVSNFGTE-VFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAI 570
SILAAWP NVPV FG++ +F+NFNL+ GTSM+CPHVAGVAALL+GAH WS ++IRSAI
Sbjct: 3 SILAAWPQNVPVEVFGSQNIFSNFNLLSGTSMACPHVAGVAALLRGAHPDWSVAAIRSAI 182
Query: 571 MTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCAL 630
MTTSD+ DNT IKD+G+ + ATP A+GAGH+NPNRALDPGLVYD+GVQDY+NLLCAL
Sbjct: 183 MTTSDMFDNTMGLIKDVGDDYKPATPLAMGAGHVNPNRALDPGLVYDVGVQDYVNLLCAL 362
Query: 631 NFTQKNISAITRSSFNDCSKPSLDLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTY 690
+TQKNI+ IT +S NDCSKPSLDLNYPSFIAF + NSS TT EF RTVTNVGE +T Y
Sbjct: 363 GYTQKNITVITGTSSNDCSKPSLDLNYPSFIAFFKS-NSSSTTQEFERTVTNVGEGQTIY 539
Query: 691 FASITPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVR 750
AS+TP+KG+ V+VIP KLVFK+KNEK SYKL+IEGP ++ VAFGYL+W D KHV+R
Sbjct: 540 VASVTPVKGYHVSVIPKKLVFKEKNEKQSYKLRIEGPIKKKEKNVAFGYLTWTDLKHVIR 719
Query: 751 SPIVVTNINFN 761
SPIVV+ + F+
Sbjct: 720 SPIVVSTLTFD 752
>TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(56%)
Length = 1773
Score = 312 bits (799), Expect = 4e-85
Identities = 194/480 (40%), Positives = 274/480 (56%), Gaps = 25/480 (5%)
Frame = +1
Query: 301 YKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGN 360
Y+D VAI F+AMEKGI VS SAGN+GP S+ N PW+ TV AGTLDR+F V LGN
Sbjct: 7 YRDSVAIGAFSAMEKGILVSCSAGNSGPGPYSLSNVAPWITTVGAGTLDRDFPAYVALGN 186
Query: 361 GVSLTGLSFYLGN-FSANNFPIVFMGMCDN-----------VKELNTVKRKIVVCEGNNE 408
G++ +G+S Y GN ++ P+V+ G N V KIV+C+
Sbjct: 187 GLNFSGVSLYRGNALPDSSLPLVYAGNVSNGAMNGNLCITGTLSPEKVAGKIVLCD-RGL 363
Query: 409 TLHEQMFNVYKAKVVGGVFISNIL----DINDVDNSFPSIIINPVNGEIVKAYIKSHNSN 464
T Q +V K+ G+ +SN ++ + P+ + G+ +K Y+ S+
Sbjct: 364 TARVQKGSVVKSAGALGMVLSNTAANGEELVADAHLLPATAVGQKAGDAIKKYLV---SD 534
Query: 465 ASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAW-----PT 519
A + F+ T G++ +P V +SSRGP++ P +LKPD+ APG +ILA W PT
Sbjct: 535 AKPTVKIFFEGTKVGIQPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAVGPT 714
Query: 520 NVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDN 579
+PV N + FN+I GTSMSCPHV+G+AAL+K AH WSP+++RSA+MTT+ +
Sbjct: 715 GLPVDNRRVD----FNIISGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTTAYTVYK 882
Query: 580 TKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISA 639
T E ++D G + +TPF G+GH++P AL+PGLVYD+ V DY+ LCALN++ IS
Sbjct: 883 TGEKLQDSATG-KPSTPFDHGSGHVDPVAALNPGLVYDLTVDDYLGFLCALNYSAAEIST 1059
Query: 640 ITRSSFN-DCSK--PSLDLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASIT- 695
+ + F D K DLNYPSF SS + + RT+TNVG TY AS+T
Sbjct: 1060LAKRKFQCDAGKQYSVTDLNYPSFAVLF---ESSGSVVKHTRTLTNVG-PAGTYKASVTS 1227
Query: 696 PIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVV 755
+++V P L F K+NEK ++ + Q + AFG + W DGKH+V SPI V
Sbjct: 1228DTASVKISVEPQVLSF-KENEKKTFTVTFSSSGSPQHTENAFGRVEWSDGKHLVGSPISV 1404
>TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(34%)
Length = 1312
Score = 228 bits (581), Expect = 8e-60
Identities = 133/323 (41%), Positives = 192/323 (59%), Gaps = 9/323 (2%)
Frame = +1
Query: 442 PSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYV 501
P+ + G+ +K Y+ S +A + F+ T G++ +P V +SSRGP++ P +
Sbjct: 85 PATAVGQKAGDAIKKYLFS---DAKPTVKILFEGTKLGIQPSPVVAAFSSRGPNSITPQI 255
Query: 502 LKPDITAPGTSILAAW-----PTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKG 556
LKPD+ APG +ILA W PT +PV N + FN+I GTSMSCPHV+G+AAL+K
Sbjct: 256 LKPDLIAPGVNILAGWSKAVGPTGLPVDNRRVD----FNIISGTSMSCPHVSGLAALIKS 423
Query: 557 AHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVY 616
AH WSP+++RSA+MTT+ + T E ++D G + +TPF G+GH++P AL+PGLVY
Sbjct: 424 AHPDWSPAAVRSALMTTAYTVYKTGEKLQDSATG-KPSTPFDHGSGHVDPVAALNPGLVY 600
Query: 617 DIGVQDYINLLCALNFTQKNISAITRSSFN-DCSK--PSLDLNYPSFIAFSNARNSSRTT 673
D+ V DY+ LCALN++ I+ + + F D K DLNYPSF + + T
Sbjct: 601 DLTVDDYLGFLCALNYSASEINTLAKRKFQCDAGKQYSVTDLNYPSFAVLFESGGVVKHT 780
Query: 674 NEFHRTVTNVGEKKTTYFASIT-PIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQK 732
RT+TNVG TY AS+T + +++V P L F K+NEK S+ + Q+
Sbjct: 781 ----RTLTNVG-PAGTYKASVTSDMASVKISVEPQVLSF-KENEKKSFTVTFSSSGSPQQ 942
Query: 733 NKVAFGYLSWRDGKHVVRSPIVV 755
AFG + W DGKHVV +PI +
Sbjct: 943 RVNAFGRVEWSDGKHVVGTPISI 1011
>TC226351 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(48%)
Length = 1490
Score = 225 bits (573), Expect = 6e-59
Identities = 146/337 (43%), Positives = 199/337 (58%), Gaps = 14/337 (4%)
Frame = +3
Query: 108 PLKLDTTHSPQFLGLNPYRGAWPTSDFGKDIIVGVIDTG-VWPESESFRDDGMTKIPSKW 166
P D+T +P +P RG+ G D+I+GV++T + ++ + + G+ +PS W
Sbjct: 531 PCSSDSTRAPT---CSPSRGS------GSDVIIGVLNTWRLAGKAGASTNTGLGPVPSTW 683
Query: 167 KGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHSNISTTILN-STRDTNGHGTHT 225
KG C+ +N+ + N CN+KLIGARFF+KG A I+ T + S RD +GHGTHT
Sbjct: 684 KGA-CE-TGTNLTASN---CNRKLIGARFFSKGVEAILGPINETEESRSARDDDGHGTHT 848
Query: 226 STTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDG 285
++TAAGS V AS FGYA+GTARG+A+ +RVA YK W G SSDI+AAI+ AI D
Sbjct: 849 ASTAAGSVVSDASLFGYASGTARGMATRARVAAYKVCW--KGGCFSSDILAAIERAILDN 1022
Query: 286 VDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAA 345
V++LS+SLG Y+D VAI F+AME GI VS SAGN GPS S+ N PW+ TV A
Sbjct: 1023VNVLSLSLGGGMSDYYRDSVAIGAFSAMENGILVSCSAGNAGPSPYSLSNVAPWITTVGA 1202
Query: 346 GTLDREFLGTVTLGNGVSLTGLSFYLGNFSANN-FPIVFMGMCDN-----------VKEL 393
GTLDR+F V LGNG++ +G+S Y GN ++ P V+ G N
Sbjct: 1203GTLDRDFPAYVALGNGLNFSGVSLYRGNAVPDSPLPFVYAGNVSNGAMNGNLCITGTLSP 1382
Query: 394 NTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISN 430
V KIV+C+ T Q +V K+ G+ +SN
Sbjct: 1383EKVAGKIVLCD-RGLTARVQKGSVVKSAGALGMVLSN 1490
Score = 77.0 bits (188), Expect = 3e-14
Identities = 42/125 (33%), Positives = 69/125 (54%), Gaps = 7/125 (5%)
Frame = +2
Query: 13 YITSLHVIFTLALSD-------NYIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNN 65
++ L V+ L L + YI+H+ S+MP+SF + WYES+L V
Sbjct: 224 FVAILWVVLFLGLHEAAEPEKSTYIVHVAKSEMPESFEHHALWYESSLKTV--------- 376
Query: 66 STSSKIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPY 125
S S++I YTY N ++G++ L+ EE L+T +G ++ +P+ +L TT +P FLGL+
Sbjct: 377 SDSAEIMYTYDNAIHGYATRLTAEEARVLETQAGILAVLPETRYELHTTRTPMFLGLDKS 556
Query: 126 RGAWP 130
+P
Sbjct: 557 ANMFP 571
>TC229906 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1,
partial (40%)
Length = 1392
Score = 223 bits (569), Expect = 2e-58
Identities = 153/454 (33%), Positives = 222/454 (48%), Gaps = 24/454 (5%)
Frame = +2
Query: 326 NGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFSANNFPIVFMG 385
+GP SI PW+++VAA T+ R+FL V LGNG+ G+S + FP+V+ G
Sbjct: 2 SGPGLSSITTYSPWILSVAASTIGRKFLTKVQLGNGMVFEGVSINTFDLKNKMFPLVYAG 181
Query: 386 MCDNVKE----------------LNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFIS 429
N + + VK KIV+C+GN + ++G
Sbjct: 182 DVPNTADGYNSSTSRFCYVNSVDKHLVKGKIVLCDGNASPKKVGDLSGAAGMLLGAT--- 352
Query: 430 NILDINDVDNSF--PSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVD 487
D+ D ++ P+ I+ N +++ +Y+ S ++ ++I TP +
Sbjct: 353 ---DVKDAPFTYALPTAFISLRNFKLIHSYMVSLRNSTATIFRSDEDNDD---SQTPFIV 514
Query: 488 FYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNF-GTEVFNNFNLIDGTSMSCPH 546
+SSRGP+ P LKPD+ APG +ILAAW +S F G + +N+ GTSM+CPH
Sbjct: 515 SFSSRGPNPLTPNTLKPDLAAPGVNILAAWSPVYTISEFKGDKRAVQYNIESGTSMACPH 694
Query: 547 VAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINP 606
V+ AA +K H WSP+ I+SA+MTT+ + T FA GAG INP
Sbjct: 695 VSAAAAYVKSFHPNWSPAMIKSALMTTATPMSPTL----------NPDAEFAYGAGLINP 844
Query: 607 NRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFNDCSKPS-----LDLNYPSFI 661
+A +PGLVYDI DY+ LC +T + + +T+ + CSK + DLN PS
Sbjct: 845 LKAANPGLVYDISEADYVKFLCGEGYTDEMLRVLTK-DHSRCSKHAKKEAVYDLNLPSLA 1021
Query: 662 AFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYK 721
+ N + SR FHRTVTNVG ++Y A + + V PN L F +K S+
Sbjct: 1022LYVNVSSFSRI---FHRTVTNVGLATSSYKAKVVSPSLIDIQVKPNVLSFTSIGQKKSFS 1192
Query: 722 LKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVV 755
+ IEG + L W DG VRSPIVV
Sbjct: 1193VIIEG---NVNPDILSASLVWDDGTFQVRSPIVV 1285
>TC229882 weakly similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like
proteinase, partial (27%)
Length = 1263
Score = 223 bits (568), Expect = 2e-58
Identities = 124/317 (39%), Positives = 181/317 (56%), Gaps = 5/317 (1%)
Frame = +1
Query: 442 PSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYV 501
P++ + + G+ ++AY +S+ + ++ F+ T VK +P V +SSRGP+ +
Sbjct: 52 PAVAVGRIVGDQIRAYA---SSDPNPTVHLDFRGTVLNVKPSPVVAAFSSRGPNMVTRQI 222
Query: 502 LKPDITAPGTSILAAWPTNVPVSNFGTEVFNN-FNLIDGTSMSCPHVAGVAALLKGAHNG 560
LKPD+ PG +ILA W + S + FN++ GTSMSCPH++G+AALLK AH
Sbjct: 223 LKPDVIGPGVNILAGWSEAIGPSGLSDDTRKTQFNIMSGTSMSCPHISGLAALLKAAHPQ 402
Query: 561 WSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGV 620
WS S+I+SA+MTT+D+ DNTK ++D G + P+A GAGH+NP++AL PGLVYD
Sbjct: 403 WSSSAIKSALMTTADVHDNTKSQLRDAA-GGAFSNPWAHGAGHVNPHKALSPGLVYDATP 579
Query: 621 QDYINLLCALNFTQKNISAITRSSFNDCSKPSLD---LNYPSFIAFSNARNSSRTTNEFH 677
DYI LC+L +T + I IT+ S +C+K D LNYPSF + R T
Sbjct: 580 SDYIKFLCSLEYTPERIQLITKRSGVNCTKRFSDPGQLNYPSFSVLFGGKRVVRYT---- 747
Query: 678 RTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKLK-IEGPRMTQKNKVA 736
R +TNVGE + Y ++ VTV P LVF K E+ Y + + +
Sbjct: 748 RVLTNVGEAGSVYNVTVDAPSTVTVTVKPAALVFGKVGERQRYTATFVSKNGVGDSVRYG 927
Query: 737 FGYLSWRDGKHVVRSPI 753
FG + W + +H VRSP+
Sbjct: 928 FGSIMWSNAQHQVRSPV 978
>TC207702 weakly similar to UP|Q8S896 (Q8S896) Subtilisin-like serine
protease AIR3 (Fragment), partial (20%)
Length = 1025
Score = 209 bits (533), Expect = 3e-54
Identities = 117/259 (45%), Positives = 158/259 (60%), Gaps = 4/259 (1%)
Frame = +2
Query: 501 VLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNF--NLIDGTSMSCPHVAGVAALLKGAH 558
+LKPD+TAPG +ILAA+ SN + F N++ GTS+SCPHVAG+A L+K H
Sbjct: 5 ILKPDVTAPGVNILAAYSELASASNLLVDNRRGFKFNVLQGTSVSCPHVAGIAGLIKTLH 184
Query: 559 NGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDI 618
WSP++I+SAIMTT+ LDNT I+D + N+ A FA G+GH+ P+ A+DPGLVYD+
Sbjct: 185 PNWSPAAIKSAIMTTATTLDNTNRPIQDAFD-NKVADAFAYGSGHVQPDLAIDPGLVYDL 361
Query: 619 GVQDYINLLCALNFTQKNISAITRSSFNDC--SKPSLDLNYPSFIAFSNARNSSRTTNEF 676
+ DY+N LCA + Q+ ISA+ + C S DLNYPS I N T
Sbjct: 362 SLADYLNFLCASGYDQQLISALNFNGTFICKGSHSVTDLNYPS-ITLPNLGLKPVTIT-- 532
Query: 677 HRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQKNKVA 736
RTVTNVG TY A++ G+ + V+P L F K EK +++ ++ +T + K
Sbjct: 533 -RTVTNVG-PPATYTANVHSPAGYTIVVVPRSLTFTKIGEKKKFQVIVQASSVTTRRKYQ 706
Query: 737 FGYLSWRDGKHVVRSPIVV 755
FG L W DGKH+VRSPI V
Sbjct: 707 FGDLRWTDGKHIVRSPITV 763
>BM143804 similar to GP|29028287|gb| subtilase {Casuarina glauca}, partial
(11%)
Length = 408
Score = 204 bits (518), Expect = 2e-52
Identities = 99/132 (75%), Positives = 113/132 (85%)
Frame = +2
Query: 279 DAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIP 338
D+AISDGVD+LS+S G DD+ LY+DPVAIATF+AMEKGIFVSTSAGN GP +HNGIP
Sbjct: 2 DSAISDGVDVLSLSFGFDDVPLYEDPVAIATFSAMEKGIFVSTSAGNEGPFLGRLHNGIP 181
Query: 339 WVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFSANNFPIVFMGMCDNVKELNTVKR 398
WVITVAAGTLDREF GT+TLGNGV +TG+S Y GNFS++N PIVFMG+CDNVKEL VK
Sbjct: 182 WVITVAAGTLDREFHGTLTLGNGVQITGMSLYHGNFSSSNVPIVFMGLCDNVKELAKVKS 361
Query: 399 KIVVCEGNNETL 410
KIVVCE N T+
Sbjct: 362 KIVVCEDKNGTI 397
>TC207663 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-like
protease) , partial (37%)
Length = 1171
Score = 202 bits (513), Expect = 6e-52
Identities = 126/304 (41%), Positives = 167/304 (54%), Gaps = 12/304 (3%)
Frame = +1
Query: 462 NSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNV 521
+S+ + A ++F T V+ +P V +SSRGP+ P +LKPD+ APG +ILA W V
Sbjct: 16 SSSPNPTAKIAFLGTHLQVQPSPVVAAFSSRGPNALTPKILKPDLIAPGVNILAGWTGAV 195
Query: 522 PVSNFGTEVFN-NFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNT 580
+ + + +FN+I GTSMSCPHV+G+AA+LKGAH WSP++IRSA+MTT+
Sbjct: 196 GPTGLTVDTRHVSFNIISGTSMSCPHVSGLAAILKGAHPQWSPAAIRSALMTTAYTSYKN 375
Query: 581 KEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAI 640
E I+DI G + TPF GAGH++P ALDPGLVYD V DY+ CALN++ I
Sbjct: 376 GETIQDISTG-QPGTPFDYGAGHVDPVAALDPGLVYDANVDDYLGFFCALNYSSFQIKLA 552
Query: 641 TRSSFNDCSKPSL---DLNYPSFI----AFSNARNSSRT--TNEFHRTVTNVGEKKTTYF 691
R + K D NYPSF S S T T ++ R +TNVG TY
Sbjct: 553 ARRDYTCDPKKDYRVEDFNYPSFAVPMDTASGIGGGSDTLKTVKYSRVLTNVG-APGTYK 729
Query: 692 ASITPI--KGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVV 749
AS+ + + V PN L F + EK Y + M +F L W DGKH V
Sbjct: 730 ASVMSLGDSNVKTVVEPNTLSFTELYEKKDYTVSFTYTSM-PSGTTSFARLEWTDGKHKV 906
Query: 750 RSPI 753
SPI
Sbjct: 907 GSPI 918
>TC229044 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, partial
(42%)
Length = 984
Score = 193 bits (491), Expect = 2e-49
Identities = 121/324 (37%), Positives = 181/324 (55%), Gaps = 8/324 (2%)
Frame = +1
Query: 441 FPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPY 500
FP+ +I+ +G + YI NS ++ +A + T K P V +SSRGPS+
Sbjct: 13 FPATVISSKDGVTILQYI---NSTSNPVATILPTATVLDYKPAPVVPNFSSRGPSSLSSN 183
Query: 501 VLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNG 560
+LKPDI APG +ILAAW N + +N+I GTSM+CPHV+G+A+ +K +
Sbjct: 184 ILKPDIAAPGVNILAAWIGNNADDVPKGRKPSLYNIISGTSMACPHVSGLASSVKTRNPT 363
Query: 561 WSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGV 620
WS S+I+SAIMT++ ++N K I + R ATP+ GAG + + +L PGLVY+
Sbjct: 364 WSASAIKSAIMTSAIQINNLKAPITT--DSGRVATPYDYGAGEMTTSESLQPGLVYETNT 537
Query: 621 QDYINLLC--ALNFTQKNISAITRSSFNDCSKPS-----LDLNYPSFIAFSNARNSSRTT 673
DY+N LC LN T + + T + C K S ++NYPS + +
Sbjct: 538 IDYLNYLCYIGLNITTVKVISRTVPANFSCPKDSSSDLISNINYPSIA----VNFTGKAA 705
Query: 674 NEFHRTVTNVGEK-KTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQK 732
RTVTNVGE+ +T Y + G +VTV P+KL F K ++K+ Y++ I +T
Sbjct: 706 VNVSRTVTNVGEEDETAYSPVVEAPSGVKVTVTPDKLQFTKSSKKLGYQV-IFSSTLTSL 882
Query: 733 NKVAFGYLSWRDGKHVVRSPIVVT 756
+ FG ++W +GK++VRSP V+T
Sbjct: 883 KEDLFGSITWSNGKYMVRSPFVLT 954
>TC228967 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-like protease
{Arabidopsis thaliana;} , partial (21%)
Length = 1106
Score = 193 bits (491), Expect = 2e-49
Identities = 123/307 (40%), Positives = 166/307 (54%), Gaps = 12/307 (3%)
Frame = +2
Query: 462 NSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAW---- 517
+S + +A + T K PS+ +SSRGP+ P +LKPDITAPG ILAAW
Sbjct: 8 HSTPNPMAQILPGTTVLETKPAPSMASFSSRGPNIVDPNILKPDITAPGVDILAAWTAED 187
Query: 518 -PTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDI 576
PT + F + +N+ GTSMSCPHVA A LLK H WS ++IRSA+MTT+
Sbjct: 188 GPTRM---TFNDKRVVKYNIFSGTSMSCPHVAAAAVLLKAIHPTWSTAAIRSALMTTAMT 358
Query: 577 LDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQK- 635
DNT + D GN ATPFA+G+GH NP RA DPGLVYD Y+ C L TQ
Sbjct: 359 TDNTGHPLTD-ETGN-PATPFAMGSGHFNPKRAADPGLVYDASYMGYLLYTCNLGVTQNF 532
Query: 636 NISAITRSSFNDCSKPSLDLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTY-FASI 694
NI+ SF + +LNYPS + T RTVTNVG ++ Y F+++
Sbjct: 533 NITYNCPKSFLE----PFELNYPSI-----QIHRLYYTKTIKRTVTNVGRGRSVYKFSAV 685
Query: 695 TPIKGFRVTVIPNKLVFKKKNEKISYKLKIEG-----PRMTQKNKVAFGYLSWRDGKHVV 749
+P K + +T PN L F +KI++ + + P +K FG+ +W H+V
Sbjct: 686 SP-KEYSITATPNILKFNHVGQKINFTITVTANWSQIPTKHGPDKYYFGWYAWTHQHHIV 862
Query: 750 RSPIVVT 756
RSP+ V+
Sbjct: 863 RSPVAVS 883
>TC232495 similar to UP|Q8W554 (Q8W554) AT3g14240/MLN21_2, partial (40%)
Length = 731
Score = 189 bits (479), Expect = 5e-48
Identities = 104/227 (45%), Positives = 143/227 (62%), Gaps = 6/227 (2%)
Frame = +3
Query: 442 PSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYV 501
P+ + G+ +++YI NS + A + FK T GV+ P V +S+RGP+ P +
Sbjct: 42 PATAVGATAGDEIRSYIG--NSRTPATATIVFKGTRLGVRPAPVVASFSARGPNPVSPEI 215
Query: 502 LKPDITAPGTSILAAWPTNVPVSNFGTEVFNN-FNLIDGTSMSCPHVAGVAALLKGAHNG 560
LKPD+ APG +ILAAWP +V S ++ FN++ GTSM+CPHV+G+AALLK AH
Sbjct: 216 LKPDVIAPGLNILAAWPDHVGPSGVPSDGRRTEFNILSGTSMACPHVSGLAALLKAAHPD 395
Query: 561 WSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGV 620
WSP+SIRSA+MTT+ +DN + I D GN ++ F GAGH++P +A++PGLVYDI
Sbjct: 396 WSPASIRSALMTTAYTVDNKGDPILDESTGN-VSSVFDYGAGHVHPVKAMNPGLVYDISS 572
Query: 621 QDYINLLCALNFTQKNISAITRSSFNDCSKP-----SLDLNYPSFIA 662
DY+N LC N+T I ITR + DCS S +LNYPS A
Sbjct: 573 NDYVNFLCNSNYTTNTIRVITRRN-ADCSGAKRAGHSGNLNYPSLSA 710
>TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(35%)
Length = 796
Score = 179 bits (454), Expect = 4e-45
Identities = 105/267 (39%), Positives = 154/267 (57%), Gaps = 14/267 (5%)
Frame = +3
Query: 316 GIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLG-NF 374
G+FVS+SAGN+GPS S+ N PW+ TV AGT+DR+F V LG+G L+G+S Y G
Sbjct: 3 GVFVSSSAGNDGPSGMSVTNLAPWLTTVGAGTIDRDFPSQVILGDGRRLSGVSLYAGAAL 182
Query: 375 SANNFPIVFMG--------MC-DNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGG 425
+ +V+ G +C +N + N VK KIV+C+ + + V KA VG
Sbjct: 183 KGKMYQLVYPGKSGILGDSLCMENSLDPNMVKGKIVICDRGSSPRVAKGLVVKKAGGVGM 362
Query: 426 VF---ISNILDINDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKS 482
+ ISN + + P+ + G+++K YI +S+ + A + FK T G+K
Sbjct: 363 ILANGISNGEGLVGDAHLLPACAVGANEGDVIKKYI---SSSTNPTATLDFKGTILGIKP 533
Query: 483 TPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEV-FNNFNLIDGTS 541
P + +S+RGP+ P +LKPD APG +ILAAW V + ++ FN++ GTS
Sbjct: 534 APVIASFSARGPNGLNPQILKPDFIAPGVNILAAWTQAVGPTGLDSDTRRTEFNILSGTS 713
Query: 542 MSCPHVAGVAALLKGAHNGWSPSSIRS 568
M+CPHV+G AALLK AH WSP+++RS
Sbjct: 714 MACPHVSGAAALLKSAHPDWSPAALRS 794
>TC221447 similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like proteinase,
partial (21%)
Length = 491
Score = 177 bits (450), Expect = 1e-44
Identities = 92/159 (57%), Positives = 112/159 (69%)
Frame = +1
Query: 214 STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSD 273
S RD +GHG+HTSTTAAGS V GAS FG+ANGTARG+A+ +R+A YK W G +SD
Sbjct: 13 SPRDDDGHGSHTSTTAAGSAVVGASLFGFANGTARGMATQARLATYKVCWL--GGCFTSD 186
Query: 274 IIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSI 333
I A ID AI DGV+ILS+S+G + YKD +AI TFAA GI VS SAGN GPS ++
Sbjct: 187 IAAGIDKAIEDGVNILSMSIGGGLMDYYKDTIAIGTFAATAHGILVSNSAGNGGPSQATL 366
Query: 334 HNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLG 372
N PW+ TV AGT+DR+F +TLGNG TG+S Y G
Sbjct: 367 SNVAPWLTTVGAGTIDRDFPAYITLGNGKMYTGVSLYNG 483
>TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-like
protease) , partial (34%)
Length = 872
Score = 176 bits (447), Expect = 3e-44
Identities = 107/260 (41%), Positives = 149/260 (57%), Gaps = 5/260 (1%)
Frame = +3
Query: 396 VKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDIND--VDNSF--PSIIINPVNG 451
V KIV+C+ E+ V A +G + +SN D + V +S+ P+ + +
Sbjct: 111 VAGKIVICDRGGNARVEKGLVVKSAGGIG-MILSNNEDYGEELVADSYLLPAAALGQKSS 287
Query: 452 EIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGT 511
+K Y+ S + + A + F T GV+ +P V +SSRGP+ P +LKPD+ APG
Sbjct: 288 NELKKYVFS---SPNPTAKLGFGGTQLGVQPSPVVAAFSSRGPNVLTPKILKPDLIAPGV 458
Query: 512 SILAAWPTNVPVSNFGTEVFN-NFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAI 570
+ILA W V + + + FN+I GTSMSCPHV G+AALLKG H WSP++IRSA+
Sbjct: 459 NILAGWTGAVGPTGLTEDTRHVEFNIISGTSMSCPHVTGLAALLKGTHPEWSPAAIRSAL 638
Query: 571 MTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCAL 630
MTT+ + IKD+ G ATPF GAGH++P A DPGLVYD V DY++ CAL
Sbjct: 639 MTTAYRTYKNGQTIKDVATG-LPATPFDYGAGHVDPVAAFDPGLVYDTTVDDYLSFFCAL 815
Query: 631 NFTQKNISAITRSSFNDCSK 650
N++ I + R F CSK
Sbjct: 816 NYSPYQIKLVARRDFT-CSK 872
>TC216757 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(20%)
Length = 1126
Score = 174 bits (441), Expect = 1e-43
Identities = 98/211 (46%), Positives = 131/211 (61%), Gaps = 6/211 (2%)
Frame = +2
Query: 554 LKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPG 613
LK AH WSP++IRSA+MTT+ +LDN + + D GN ++TP+ GAGH+N RA+DPG
Sbjct: 2 LKSAHPDWSPAAIRSAMMTTATVLDNRNKTMTDEATGN-SSTPYDFGAGHLNLGRAMDPG 178
Query: 614 LVYDIGVQDYINLLCALNFTQKNISAITRSSFN-DCSKPSLD-LNYPSFIAFSNARNSSR 671
LVYDI DY+N LC + + K I ITR+ + +P+ + LNYPSF+A A +
Sbjct: 179 LVYDITNNDYVNFLCGIGYGPKVIQVITRAPASCPVRRPAPENLNYPSFVAMFPASSKGV 358
Query: 672 TTNEFHRTVTNVGEKKTTYFASI-TPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMT 730
+ F RTVTNVG + Y S+ P G VTV P++LVF + +K SY + + G
Sbjct: 359 ASKTFIRTVTNVGPANSVYRVSVEAPASGVSVTVKPSRLVFSEAVKKRSYVVTVAGDTRK 538
Query: 731 QK---NKVAFGYLSWRDGKHVVRSPIVVTNI 758
K + FG L+W DGKHVVRSPIVVT I
Sbjct: 539 LKMGPSGAVFGSLTWTDGKHVVRSPIVVTQI 631
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,633,317
Number of Sequences: 63676
Number of extensions: 449054
Number of successful extensions: 2913
Number of sequences better than 10.0: 125
Number of HSP's better than 10.0 without gapping: 2690
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2729
length of query: 762
length of database: 12,639,632
effective HSP length: 105
effective length of query: 657
effective length of database: 5,953,652
effective search space: 3911549364
effective search space used: 3911549364
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144727.4


