

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144726.1 + phase: 0
(240 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
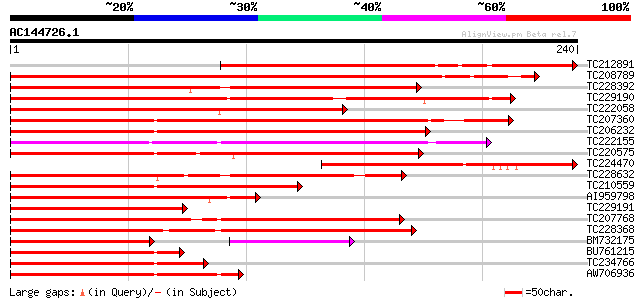
Score E
Sequences producing significant alignments: (bits) Value
TC212891 homologue to UP|Q9AR13 (Q9AR13) MADS-box transcription ... 260 3e-70
TC208789 similar to UP|Q9SBQ1 (Q9SBQ1) MADS box transcription fa... 258 2e-69
TC228392 similar to UP|Q8LLR0 (Q8LLR0) MADS-box protein 4, parti... 185 2e-47
TC229190 similar to UP|Q9SEG4 (Q9SEG4) CAGL2, partial (86%) 177 4e-45
TC222058 homologue to UP|AGL9_PETHY (Q03489) Agamous-like MADS b... 165 2e-41
TC207360 similar to UP|Q8VWZ3 (Q8VWZ3) C-type MADS box protein, ... 160 5e-40
TC206232 homologue to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS... 145 1e-35
TC222155 weakly similar to UP|O81662 (O81662) Transcription acti... 139 9e-34
TC220575 similar to UP|Q8LLR1 (Q8LLR1) MADS-box protein 3, parti... 138 2e-33
TC224470 similar to UP|Q9AR13 (Q9AR13) MADS-box transcription fa... 134 3e-32
TC228632 similar to UP|Q9ZTQ9 (Q9ZTQ9) MADS box protein 26, part... 125 1e-29
TC210559 similar to UP|Q84MI1 (Q84MI1) MADS-box protein (Fragmen... 120 6e-28
AI959798 similar to GP|22091481|emb MADS box transcription facto... 119 9e-28
TC229191 homologue to UP|Q84U54 (Q84U54) MADS-RIN-like protein, ... 116 8e-27
TC207768 similar to UP|Q9ST53 (Q9ST53) MADS-box protein 4, parti... 112 2e-25
TC228368 similar to UP|O49173 (O49173) MADS-box protein NMH 7, p... 111 3e-25
BM732175 similar to GP|15022157|gb MADS box protein-like protein... 84 6e-24
BU761215 homologue to PIR|T08039|T08 MADS-box protein - cucumber... 100 6e-22
TC234766 homologue to UP|Q7Y137 (Q7Y137) MADS-box protein PTM5, ... 98 3e-21
AW706936 97 5e-21
>TC212891 homologue to UP|Q9AR13 (Q9AR13) MADS-box transcription factor
MADS4, partial (57%)
Length = 688
Score = 260 bits (665), Expect = 3e-70
Identities = 134/151 (88%), Positives = 141/151 (92%)
Frame = +3
Query: 90 NWTIEYTRLKAKIDLLQRNYRHYMGEDLGSMSLKELQSLEQQLDTALKLIRTRRNQVMYE 149
NWTIEYTRLKAKIDLLQRN+RHYMGEDLGSMSLKELQSLEQQLDTALK IRTRRNQ+MYE
Sbjct: 3 NWTIEYTRLKAKIDLLQRNHRHYMGEDLGSMSLKELQSLEQQLDTALKQIRTRRNQLMYE 182
Query: 150 SISELQKKEKVIQEQNNMLAKKIKEKEKIAAQQQAQWEHPNHHGVNPNYLLQQQLPTLNM 209
SISELQKKEKVIQEQNNMLAKKIKEKEK+AA QQAQWEHPN GVN ++LL Q P LNM
Sbjct: 183 SISELQKKEKVIQEQNNMLAKKIKEKEKVAA-QQAQWEHPN-PGVNASFLLPQ--PLLNM 350
Query: 210 GGNYREEAPEMGRNELDLTLEPLYTCHLGCF 240
GGNYREEA E+GRNELDLTLEPLY+CHLGCF
Sbjct: 351 GGNYREEASEVGRNELDLTLEPLYSCHLGCF 443
>TC208789 similar to UP|Q9SBQ1 (Q9SBQ1) MADS box transcription factor,
partial (72%)
Length = 995
Score = 258 bits (659), Expect = 2e-69
Identities = 139/224 (62%), Positives = 175/224 (78%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKI QVTFSKRR+GLLKKA EISVLCDA+VALIVFS KGKL +Y+
Sbjct: 219 MGRGRVELKRIENKIFMQVTFSKRRSGLLKKAREISVLCDADVALIVFSTKGKLLDYSNQ 398
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
C E+ILERYERYSYAERQLV +D NW IE+ +LKA++++LQRN R++MGEDL S+
Sbjct: 399 PCTERILERYERYSYAERQLVGDDQPPNENWVIEHEKLKARVEVLQRNQRNFMGEDLDSL 578
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAA 180
+L+ LQSLEQQLD+ALK IR+R+NQ M ESISELQKK++ ++E NN+L+KKIKEKEK
Sbjct: 579 NLRGLQSLEQQLDSALKHIRSRKNQAMNESISELQKKDRTLREHNNLLSKKIKEKEKELT 758
Query: 181 QQQAQWEHPNHHGVNPNYLLQQQLPTLNMGGNYREEAPEMGRNE 224
Q+ Q N+ V+ + L+ Q L +L +GG +PE+ NE
Sbjct: 759 PQE-QEGLQNNMDVS-SVLVTQPLESLTIGG-----SPEVKSNE 869
>TC228392 similar to UP|Q8LLR0 (Q8LLR0) MADS-box protein 4, partial (98%)
Length = 755
Score = 185 bits (469), Expect = 2e-47
Identities = 99/181 (54%), Positives = 135/181 (73%), Gaps = 7/181 (3%)
Frame = +1
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVALI+FS++GK +E+ +
Sbjct: 40 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKQYEFCSG 219
Query: 61 SCMEKILERYERYSY-------AERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYM 113
S M K LERY++ +Y A ++ + + SQ EY RLKA+ + LQR+ R+ M
Sbjct: 220 SSMLKTLERYQKCNYGAPEDNVATKEALVLELSSQQ----EYLRLKARYEALQRSQRNLM 387
Query: 114 GEDLGSMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIK 173
GEDLG +S KEL+SLE+QLD++LK IR+ R Q M + +S+LQ+KE + E N L ++++
Sbjct: 388 GEDLGPLSSKELESLERQLDSSLKQIRSIRTQFMLDQLSDLQRKEHFLGESNRDLRQRLE 567
Query: 174 E 174
E
Sbjct: 568 E 570
>TC229190 similar to UP|Q9SEG4 (Q9SEG4) CAGL2, partial (86%)
Length = 1062
Score = 177 bits (449), Expect = 4e-45
Identities = 101/222 (45%), Positives = 145/222 (64%), Gaps = 8/222 (3%)
Frame = +1
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVALI+FS +GKL+E+ +
Sbjct: 313 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 492
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
+ M K LERY++ SY ++ E + ++ EY +LKA+ + LQR R+ +GEDLG +
Sbjct: 493 NSMLKTLERYQKCSYGAVEVSKPGKELESSYR-EYLKLKARFESLQRTQRNLLGEDLGPL 669
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKE------ 174
+ K+L+ LE+QLD++LK + Q M + +++LQ KE ++ E N L K++E
Sbjct: 670 NTKDLEQLERQLDSSLK-----QTQFMLDQLADLQNKEHMLVEANRSLTMKLEEINSRNQ 834
Query: 175 --KEKIAAQQQAQWEHPNHHGVNPNYLLQQQLPTLNMGGNYR 214
+ A +Q + N H L+ PTL +G +YR
Sbjct: 835 YRQTWEAGEQSMSYGTQNAHSQGFFQPLECN-PTLQIGSDYR 957
>TC222058 homologue to UP|AGL9_PETHY (Q03489) Agamous-like MADS box protein
AGL9 homolog (Floral homeotic protein FBP2) (Floral
binding protein 2), partial (60%)
Length = 495
Score = 165 bits (418), Expect = 2e-41
Identities = 86/144 (59%), Positives = 115/144 (79%), Gaps = 1/144 (0%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVALI+FS++GKL+E+ +
Sbjct: 60 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 239
Query: 61 SCMEKILERYERYSYAERQLVANDSES-QGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGS 119
S M K LERY++ +Y + + E+ + + EY +LKA+ + LQR+ R+ MGEDLG
Sbjct: 240 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 419
Query: 120 MSLKELQSLEQQLDTALKLIRTRR 143
+S KEL+SLE+QLD++LK IR+ R
Sbjct: 420 LSSKELESLERQLDSSLKQIRSTR 491
>TC207360 similar to UP|Q8VWZ3 (Q8VWZ3) C-type MADS box protein, partial
(88%)
Length = 1075
Score = 160 bits (405), Expect = 5e-40
Identities = 90/213 (42%), Positives = 139/213 (65%)
Frame = +2
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++++KRIEN NRQVTF KRR GLLKKA+E+SVLCDAEVAL+VFS +G+L+EYA +
Sbjct: 95 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSTRGRLYEYANN 274
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S + +ERY++ + A + + + E ++L+ +I +Q RH +GE LGS+
Sbjct: 275 S-VRATIERYKKANAAASNAESVSEANTQFYQQESSKLRRQIRDIQNLNRHILGEALGSL 451
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAA 180
SLKEL++LE +L+ L +R+R+++ ++ + +QK+E +Q NN L KI E E+ A
Sbjct: 452 SLKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKREIELQNHNNYLRAKIAEHER-AQ 628
Query: 181 QQQAQWEHPNHHGVNPNYLLQQQLPTLNMGGNY 213
QQQ+ +N + L + LP+ + N+
Sbjct: 629 QQQS--------NMNMSGTLCESLPSQSYDRNF 703
>TC206232 homologue to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS-2, partial
(95%)
Length = 960
Score = 145 bits (367), Expect = 1e-35
Identities = 74/178 (41%), Positives = 119/178 (66%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++++KRIEN NRQVTF KRR GLLKKA+E+SVLCDAEVALIVFS +G+L+EY+ +
Sbjct: 75 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSNN 254
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
+ + +ERY++ + + E +L+ +I +LQ + RH MG+ L ++
Sbjct: 255 N-IRSTIERYKKACSDHSSASTTTEINAQYYQQESAKLRQQIQMLQNSNRHLMGDALSTL 431
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKI 178
++KEL+ LE +L+ + IR+++++++ I QK+E ++ +N L KI + E+I
Sbjct: 432 TVKELKQLENRLERGITRIRSKKHEMLLAEIEYFQKREIELENENLCLRTKITDVERI 605
>TC222155 weakly similar to UP|O81662 (O81662) Transcription activator,
partial (65%)
Length = 915
Score = 139 bits (351), Expect = 9e-34
Identities = 82/204 (40%), Positives = 122/204 (59%)
Frame = +2
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M RG+VQLK+IE+ +RQV FSKRR+GLLKKA+E+SVLCDAEVA+IVFS G+L+E+++
Sbjct: 152 MARGKVQLKKIEDTTSRQVAFSKRRSGLLKKAYELSVLCDAEVAVIVFSQNGRLYEFSS- 328
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S M KILERY ++ D Q ++ L KI+LL+ + R +G+ + S
Sbjct: 329 SDMTKILERYREHTKDVPASKFGDDYIQ-QLKLDSASLAKKIELLEHSKRELLGQSVSSC 505
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAA 180
S EL+ +E+QL +L+ +R R+ Q+ E I +L+ +E + ++N L+ + EK
Sbjct: 506 SYDELKGIEEQLQISLQRVRQRKTQLYTEQIDQLRSQESNLLKENAKLSAMWQRAEK--- 676
Query: 181 QQQAQWEHPNHHGVNPNYLLQQQL 204
Q QW P+ Q L
Sbjct: 677 SSQQQWPRHTQAEAEPHCSSSQSL 748
>TC220575 similar to UP|Q8LLR1 (Q8LLR1) MADS-box protein 3, partial (44%)
Length = 851
Score = 138 bits (348), Expect = 2e-33
Identities = 80/177 (45%), Positives = 114/177 (64%), Gaps = 2/177 (1%)
Frame = +2
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG+V L+RI+NKINRQVTFSKRR GLLKKA E+SVLCDAE+AL++FS +GKLF+Y++
Sbjct: 158 MGRGKVVLERIQNKINRQVTFSKRRNGLLKKAFELSVLCDAEIALVIFSSRGKLFQYSST 337
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTI--EYTRLKAKIDLLQRNYRHYMGEDLG 118
+ +I+E+Y + + Q + +E Q + E L+ K + LQR R+ +GE+L
Sbjct: 338 D-INRIIEKYRQCCFNMSQ-TGDVAEHQSEQCLYQELLILRVKHESLQRTQRNLLGEELE 511
Query: 119 SMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEK 175
+S+KEL SLE+QLD L R Q + I EL K +++ N L + + K
Sbjct: 512 PLSMKELHSLEKQLDRTLGQARKHLTQKLISRIDELHGKVHNLEQVNKHLESQERGK 682
>TC224470 similar to UP|Q9AR13 (Q9AR13) MADS-box transcription factor MADS4,
partial (33%)
Length = 485
Score = 134 bits (338), Expect = 3e-32
Identities = 75/112 (66%), Positives = 90/112 (79%), Gaps = 4/112 (3%)
Frame = +2
Query: 133 DTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAAQQQAQWEHPNHH 192
+ +K IRTRRN +M ESISELQKKEK IQE+NN LAKKIKEKE+ AQQ AQWE PN+
Sbjct: 5 EAGIKNIRTRRNDLMSESISELQKKEKRIQEENNTLAKKIKEKEQAVAQQAAQWEQPNYR 184
Query: 193 GVNPNYLLQQQ-LPT-LNM-GGNY-REEAPEMGRNELDLTLEPLYTCHLGCF 240
V+ +++ QQQ LPT LN+ G NY +E APE+GRN LDLTLEPLY+C+LGCF
Sbjct: 185 -VDTSFMPQQQPLPTSLNICGNNYCQEAAPELGRNGLDLTLEPLYSCNLGCF 337
>TC228632 similar to UP|Q9ZTQ9 (Q9ZTQ9) MADS box protein 26, partial (82%)
Length = 880
Score = 125 bits (315), Expect = 1e-29
Identities = 72/169 (42%), Positives = 108/169 (63%), Gaps = 1/169 (0%)
Frame = +1
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++++KRIEN NRQVT+SKR+ G+LKKA EI+VLCDA+V+LI+F+ GK+ +Y +
Sbjct: 55 MGRGKIEIKRIENSSNRQVTYSKRKNGILKKAKEITVLCDAQVSLIIFAASGKMHDYISP 234
Query: 61 S-CMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGS 119
S + ILERY + S +R A G E RLK + D +Q RH G+D+ S
Sbjct: 235 STTLIDILERYHKTS-GKRLWDAKHENLNG----EIERLKKENDSMQIELRHLKGDDINS 399
Query: 120 MSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNML 168
++ KEL +LE L+T L +R ++ V L++ +K+++E+N L
Sbjct: 400 LNYKELMALEDALETGLVSVREKQMDV----YRMLRRNDKILEEENREL 534
>TC210559 similar to UP|Q84MI1 (Q84MI1) MADS-box protein (Fragment), partial
(64%)
Length = 710
Score = 120 bits (301), Expect = 6e-28
Identities = 64/124 (51%), Positives = 89/124 (71%)
Frame = +1
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M RG Q++RIEN +RQVTFSKRR GLLKKA E+SVLCDAEVALI+FS +GKL+E+A+
Sbjct: 340 MVRGETQMRRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEFASS 519
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S M+ +ERY R++ + + + +D ++ + E L KI+LL+ + R +GE LGS
Sbjct: 520 S-MQDTIERYRRHNRSAQTVNRSDEQNMQHLKQETANLMKKIELLEASKRKLLGEGLGSC 696
Query: 121 SLKE 124
SL+E
Sbjct: 697 SLEE 708
>AI959798 similar to GP|22091481|emb MADS box transcription factor {Daucus
carota subsp. sativus}, partial (39%)
Length = 409
Score = 119 bits (299), Expect = 9e-28
Identities = 62/107 (57%), Positives = 83/107 (76%), Gaps = 1/107 (0%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG+V+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVALI+FS++GKL+E+ +
Sbjct: 90 MGRGKVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSG 269
Query: 61 SCMEKILERYERYSYAERQLVAN-DSESQGNWTIEYTRLKAKIDLLQ 106
K LERY R SY ++ + E+Q + EY +LK++++ LQ
Sbjct: 270 HSTAKTLERYHRCSYGALEVQHQPEIETQRRYQ-EYLKLKSRVEALQ 407
>TC229191 homologue to UP|Q84U54 (Q84U54) MADS-RIN-like protein, partial
(32%)
Length = 558
Score = 116 bits (291), Expect = 8e-27
Identities = 56/75 (74%), Positives = 68/75 (90%)
Frame = +2
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVALI+FS++GKL+E+ +
Sbjct: 320 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 499
Query: 61 SCMEKILERYERYSY 75
S M K LERY++ SY
Sbjct: 500 SSMLKTLERYQKCSY 544
>TC207768 similar to UP|Q9ST53 (Q9ST53) MADS-box protein 4, partial (27%)
Length = 951
Score = 112 bits (279), Expect = 2e-25
Identities = 62/167 (37%), Positives = 103/167 (61%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MG+ +V++KRIENK RQ+TFSKRR GL+KKA E+S+LCDA+VAL++FS GKL+E
Sbjct: 111 MGKKKVEIKRIENKSTRQITFSKRRNGLMKKARELSILCDAKVALLIFSSTGKLYELCNG 290
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
+ +++++Y + A D++SQ E + + Q RH+ +L +
Sbjct: 291 DSLAEVVQQYWDHLGAS----GTDTKSQ-ELCFEIADIWSGSAFSQMIKRHFGVSELEHL 455
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNM 167
S+ +L LE+ AL IR+ + ++M ES+ L+KK + +++ N++
Sbjct: 456 SVSDLMELEKLTHAALSRIRSAKMRLMMESVVNLKKKIEALEKTNDV 596
>TC228368 similar to UP|O49173 (O49173) MADS-box protein NMH 7, partial (97%)
Length = 1042
Score = 111 bits (277), Expect = 3e-25
Identities = 64/172 (37%), Positives = 104/172 (60%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M RG++Q+KRIEN NRQVT+SKRR GL KKA+E++VLCDA+V++I+FS GKL +Y +
Sbjct: 81 MARGKIQIKRIENNTNRQVTYSKRRNGLFKKANELTVLCDAKVSIIMFSSTGKLHQYISP 260
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S K + +++Y + N N +LK L++ R MG+ L +
Sbjct: 261 STSTK--QFFDQYQMTLGVDLWNSHYE--NMQENLKKLKEVNRNLRKEIRQRMGDCLNEL 428
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKI 172
+++L+ LE+++D A K++R R+ +V+ I +KK +E +N L + +
Sbjct: 429 GMEDLKLLEEEMDKAAKVVRERKYKVITNQIDTQRKKFNNEKEVHNRLLRDL 584
>BM732175 similar to GP|15022157|gb MADS box protein-like protein NGL9
{Medicago sativa}, partial (60%)
Length = 421
Score = 83.6 bits (205), Expect(2) = 6e-24
Identities = 37/61 (60%), Positives = 52/61 (84%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++++KRIEN NRQVT+SKR+ G+LKKA EI+VLCDA+V+LI+F+ GK+ +Y +
Sbjct: 39 MGRGKIEIKRIENSSNRQVTYSKRKNGILKKAKEITVLCDAQVSLIIFAASGKMHDYISP 218
Query: 61 S 61
S
Sbjct: 219S 221
Score = 44.3 bits (103), Expect(2) = 6e-24
Identities = 22/53 (41%), Positives = 32/53 (59%)
Frame = +2
Query: 94 EYTRLKAKIDLLQRNYRHYMGEDLGSMSLKELQSLEQQLDTALKLIRTRRNQV 146
E RLK + D +Q RH GED+ S++ KEL +LE L+T L +R ++ V
Sbjct: 239 EIERLKKENDSMQIELRHLKGEDINSLNYKELMALEDALETGLVSVREKQMDV 397
>BU761215 homologue to PIR|T08039|T08 MADS-box protein - cucumber, partial
(42%)
Length = 450
Score = 100 bits (249), Expect = 6e-22
Identities = 49/74 (66%), Positives = 64/74 (86%)
Frame = +1
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++++KRIEN NRQVTF KRR GLLKKA+E+SVLCDAEVALIVFS +G+L+EYA +
Sbjct: 142 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANN 321
Query: 61 SCMEKILERYERYS 74
S ++ +ERY++ S
Sbjct: 322 S-VKATIERYKKAS 360
>TC234766 homologue to UP|Q7Y137 (Q7Y137) MADS-box protein PTM5, partial
(33%)
Length = 490
Score = 98.2 bits (243), Expect = 3e-21
Identities = 50/84 (59%), Positives = 67/84 (79%)
Frame = +2
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M RG+ QL+RIEN +RQVTFSKRR GLLKKA E+SVLCDAEVALI+FS +GKL+E+A+
Sbjct: 239 MVRGKTQLRRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEFASS 418
Query: 61 SCMEKILERYERYSYAERQLVAND 84
S M+ +ERY R++ + + + +D
Sbjct: 419 S-MQDTIERYRRHNRSAQTVNRSD 487
>AW706936
Length = 422
Score = 97.4 bits (241), Expect = 5e-21
Identities = 50/99 (50%), Positives = 70/99 (70%)
Frame = +2
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++ ++RI+N +RQVTFSKRR GLLKKA E+S+LCDAEV L+VFS GKL++YA+
Sbjct: 131 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKARELSILCDAEVGLMVFSSTGKLYDYAST 310
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLK 99
S M+ ++ERY + L+ SE++ W E L+
Sbjct: 311 S-MKSVIERYNKLKEEHHHLMNPASEAK-FWQTEAASLR 421
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,849,255
Number of Sequences: 63676
Number of extensions: 98190
Number of successful extensions: 700
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 681
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 686
length of query: 240
length of database: 12,639,632
effective HSP length: 94
effective length of query: 146
effective length of database: 6,654,088
effective search space: 971496848
effective search space used: 971496848
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC144726.1


