

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144724.7 - phase: 0 /pseudo
(1419 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
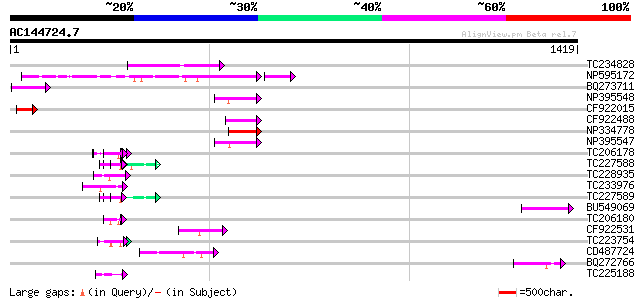
Score E
Sequences producing significant alignments: (bits) Value
TC234828 205 1e-52
NP595172 polyprotein [Glycine max] 170 4e-43
BQ273711 77 7e-14
NP395548 reverse transcriptase [Glycine max] 74 6e-13
CF922015 73 1e-12
CF922488 72 1e-12
NP334778 reverse transcriptase [Glycine max] 69 1e-11
NP395547 reverse transcriptase [Glycine max] 69 1e-11
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 60 5e-09
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 59 1e-08
TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana geno... 58 3e-08
TC233976 57 6e-08
TC227589 57 6e-08
BU549069 53 8e-07
TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 49 1e-05
CF922531 47 8e-05
TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequen... 46 1e-04
CD487724 46 1e-04
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 46 1e-04
TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like prot... 45 3e-04
>TC234828
Length = 857
Score = 205 bits (522), Expect = 1e-52
Identities = 113/252 (44%), Positives = 148/252 (57%), Gaps = 8/252 (3%)
Frame = +3
Query: 294 KAAGKVFALNAEEVEQPDNLIRGMCFINSTHLIGIIDIGATHSFISVSCVKRLKLVVTPL 353
K +VFA++ E D+LIRG C I L + D GATHSFIS +CV+RL L T L
Sbjct: 120 KVPSRVFAMSGSEAAASDDLIRGKCLIADKLLDVLYDSGATHSFISHACVERLGLCATEL 299
Query: 354 LRGMVIDTPARGSVTTSFMCAKCPVNFGNVDFELDLVCLPLKHMDVIFGMDWLLFFGVSI 413
MV+ TP VTTS +C KCP+ F DL+CLPL H+DVI GMDWL + +
Sbjct: 300 PYDMVVSTPTSEPVTTSRVCLKCPIIVEGRSFMADLICLPLAHLDVILGMDWLSTNHIFL 479
Query: 414 NCLTKSVTFSKPVEKLDRKFLTAEQVKKSLDGEAC----VFMMFASLKENSEKGVGDLPI 469
+C K + F V + +E +K+ E +M+ S+ + V +P+
Sbjct: 480 DCKEKMLVFGGDV-------VPSEPLKEDAANEETEDVRTYMVLFSMYVEEDAEVSCIPV 638
Query: 470 VQEFPE----DITELPLEREVEFAIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQ 525
V EFPE D+ ELP EREVEF ID+VPG +P+ I PY MS EL E+K Q+++LL KQ
Sbjct: 639 VSEFPEVFPDDVCELPPEREVEFIIDVVPGANPVSIAPYRMSPVELAEVKAQVQDLLSKQ 818
Query: 526 FIRPSVSPWGAP 537
F+RPS SPWGAP
Sbjct: 819 FVRPSASPWGAP 854
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 170 bits (430), Expect(2) = 4e-43
Identities = 171/666 (25%), Positives = 280/666 (41%), Gaps = 66/666 (9%)
Frame = +1
Query: 31 RFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWWTNAKGRLEIVGEVVTW 90
RFD + W+ E+ F T D ++ + + L ++ W+ L+ +W
Sbjct: 310 RFDGKNVMDWIFKAEQFFDYYATPDADRLIIASVHLDQDVVPWYQ----MLQKTEPFSSW 477
Query: 91 AKFKAEFLRKYFPEDLRTRKEVAF-LNLK*GSISVAEYAAKFEKLSRFCPYIIAEDAMVS 149
F + P + F LN S +V EY +F L + AE +
Sbjct: 478 QAFTRALELDFGPSAYDCPRATLFKLNQ---SATVNEYYMQFTALVNRVDGLSAE--AIL 642
Query: 150 KCVKFESGLRPDIYQYMCVQEIRDFDTLVHKCRKFDD--AGRVKANYYKAQSEKRGKGHG 207
C F SGL+ +I + + E R V + F++ K + +
Sbjct: 643 DC--FVSGLQEEISRDVKAMEPRTLTKAVALAKLFEEKYTSPPKTKTFSNLARNFTSNTS 816
Query: 208 VGKPYNKDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAEC 267
+ Y +K KP+L + + N K IS AE
Sbjct: 817 ATQKYPPTNQKN----DNPKPNLPPLLPTPSTKPFNLRNQNIKKIS----------PAEI 954
Query: 268 KDVARDVTCYNCGEKGHISTKCTKPKKAAGKVFALNAEEVEQPD---------------- 311
+ CY C EK + KC P + +V L EE ++
Sbjct: 955 QLRREKNLCYFCDEKFSPAHKC--PNR---QVMLLQLEETDEDQTDEQVMVTEEANMDDD 1119
Query: 312 ------NLIRGMCFINSTHLIG---------IIDIGATHSFISVSCVKRLKLVVTPL--L 354
N +RG + + G ++D G++ +FI + LKL V P L
Sbjct: 1120THHLSLNAMRGSNGVGTIRFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKLPVEPAPNL 1299
Query: 355 RGMVIDTPARGSVTTSF-MCAKCPVNFGNVDFELDLVCLPLKHMDVIFGMDWLLFFGVSI 413
R +V G + ++ + + P++ + ++ + L + DVI G WL G +
Sbjct: 1300RVLV----GNGQILSAEGIVQQLPLHIQGQEVKVPVYLLQISGADVILGSTWLATLGPHV 1467
Query: 414 -NCLTKSVTFSKPVEKLDRKFLTAE----------------QVKKSLDGEACVFMMFASL 456
+ ++ F + + KF+T + +++ + E C F +
Sbjct: 1468ADYAALTLKFFQ-----NDKFITLQGEGNSEATQAQLHHFRRLQNTKSIEEC-FAIQLIQ 1629
Query: 457 KENSEKGVGDLP---------IVQEFPEDI---TELPLEREVEFAIDLVPGMSPIWITPY 504
KE E + DLP ++ + + LP +RE + AI L G P+ + PY
Sbjct: 1630KEVPEDTLKDLPTNIDPELAILLHTYAQVFAVPASLPPQREQDHAIPLKQGSGPVKVRPY 1809
Query: 505 PMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKN 564
++ +++K ++E+L + I+PS SP+ P+LLVKKKDGS R C DYR LN +T+K+
Sbjct: 1810RYPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKD 1989
Query: 565 RYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGV 624
+P+ +D+L+D+L GA+ FSK+D RSGYH V+ ED KT FRT +GHYE+ VM FG+
Sbjct: 1990SFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGL 2169
Query: 625 YNVPGT 630
N P T
Sbjct: 2170TNAPAT 2187
Score = 25.0 bits (53), Expect(2) = 4e-43
Identities = 23/79 (29%), Positives = 34/79 (42%)
Frame = +2
Query: 637 EYFILIWTVLWWYL*MLFWCIRNLRRSMLSI*GLCCKY*RRTSYVLSCLRVSFGCRK*VS 696
+YF L+ L+ M I L R + S LC + * SY+L C V +K ++
Sbjct: 2207 KYFSLLSENLFLCFLMTS*YIVLLGRIISSTWNLCYRP*SNISYLLDCPNVHLEIQKLIT 2386
Query: 697 LVM*FLRVVFSWIRRRLMQ 715
+L F W +R Q
Sbjct: 2387 WATRYLG*EFPWRIQRYKQ 2443
>BQ273711
Length = 409
Score = 76.6 bits (187), Expect = 7e-14
Identities = 39/98 (39%), Positives = 52/98 (52%)
Frame = -2
Query: 4 IGHRSEGKVEDRRLDRFLRNNPPTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTT 63
+ R E R L F +N+PP ++G +DPEGA+ W+ E+IF AM ++ KV T
Sbjct: 294 VQERDVEPAEYRGLMAFRKNHPPKFSGDYDPEGARLWLAETEKIFEAMGCLEEHKVLYAT 115
Query: 64 HMLAEEAEYWWTNAKGRLEIVGEVVTWAKFKAEFLRKY 101
ML EAE WW K G V+ W FK +FL Y
Sbjct: 114 FMLQGEAENWWKFVKPSFVAPGGVIPWNAFKDKFLENY 1
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 73.6 bits (179), Expect = 6e-13
Identities = 45/136 (33%), Positives = 72/136 (52%), Gaps = 19/136 (13%)
Frame = +1
Query: 514 LKKQLEELLEKQFIRP-SVSPWGAPVLLVKKKDG------------------SVRLCVDY 554
++K++ +LLE I P S S W +PVL+V KK+G S +LC+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 555 RQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGH 614
R+LN+ T K+ +PL +D ++++L G + +D GY+ V +D K F +G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 615 YEYFVMQFGVYNVPGT 630
+ Y + FG+ N P T
Sbjct: 361 FAYRRIPFGLCNAPTT 408
>CF922015
Length = 172
Score = 72.8 bits (177), Expect = 1e-12
Identities = 32/54 (59%), Positives = 40/54 (73%)
Frame = -3
Query: 17 LDRFLRNNPPTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEA 70
LDRF RNNPPT+ G +DPEGA+ W++ +E+IF M D QKV THMLA+EA
Sbjct: 164 LDRFQRNNPPTFKGGYDPEGAEAWLREIEKIFRVMECQDHQKVLFATHMLADEA 3
>CF922488
Length = 741
Score = 72.4 bits (176), Expect = 1e-12
Identities = 38/90 (42%), Positives = 54/90 (59%)
Frame = +3
Query: 541 VKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKA 600
V K+DG V +CVDYR LN + K+++PL I+ L+D FS +D SGY+ ++
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 601 EDISKTVFRTRYGHYEYFVMQFGVYNVPGT 630
ED+ KT F T +G + Y M FG+ NV T
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGAT 272
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 69.3 bits (168), Expect = 1e-11
Identities = 35/82 (42%), Positives = 50/82 (60%)
Frame = +3
Query: 549 RLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVF 608
R+CVDYR LN+ + K+ +PL ID LM + +FS +D SGY+ ++ ED+ KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 609 RTRYGHYEYFVMQFGVYNVPGT 630
T +G + Y VM FG+ N T
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGAT 248
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 69.3 bits (168), Expect = 1e-11
Identities = 44/136 (32%), Positives = 67/136 (48%), Gaps = 19/136 (13%)
Frame = +1
Query: 514 LKKQLEELLEKQFIRP-SVSPWGAPVLLVKKKDGSV------------------RLCVDY 554
++K++ +LLE I P S S W +PV +V KK G R+C+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 555 RQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGH 614
R+LN+ T K+ YPL +D ++ +L + +D SGY+ V +D KT F +
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 615 YEYFVMQFGVYNVPGT 630
+ Y M FG+ N T
Sbjct: 361 FAYRRMPFGLCNASTT 408
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 60.5 bits (145), Expect = 5e-09
Identities = 30/79 (37%), Positives = 41/79 (50%)
Frame = +2
Query: 211 PYNKDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDV 270
PY +D R+ G S+ +L C C GHYA +C CH CG GH A+EC
Sbjct: 299 PYRRDSRR-----GFSRDNL----CKNCKRPGHYARECPNVAICHNCGLPGHIASECTTK 451
Query: 271 ARDVTCYNCGEKGHISTKC 289
+ C+NC E GH+++ C
Sbjct: 452 S---LCWNCKEPGHMASSC 499
Score = 59.7 bits (143), Expect = 9e-09
Identities = 26/63 (41%), Positives = 34/63 (53%), Gaps = 4/63 (6%)
Frame = +2
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARDV----TCYNCGEKGHISTKCT 290
C+ C GH A+ C + CH CGKAGH+A EC C NC ++GHI+ +CT
Sbjct: 458 CWNCKEPGHMASSCPNEGICHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAECT 637
Query: 291 KPK 293
K
Sbjct: 638 NEK 646
Score = 53.1 bits (126), Expect = 8e-07
Identities = 22/69 (31%), Positives = 35/69 (49%)
Frame = +2
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCTKPKK 294
C+ CG GH A++C C C + GH A+ C + + C+ CG+ GH + +C+ P
Sbjct: 401 CHNCGLPGHIASECTTKSLCWNCKEPGHMASSCPN---EGICHTCGKAGHRARECSAPPM 571
Query: 295 AAGKVFALN 303
G + N
Sbjct: 572 PPGDLRLCN 598
Score = 48.9 bits (115), Expect = 2e-05
Identities = 26/86 (30%), Positives = 39/86 (45%), Gaps = 1/86 (1%)
Frame = +2
Query: 207 GVGKPYNKDKRKKREVGGGSKPSLADIK-CYRCGTLGHYANDCKKDISCHKCGKAGHKAA 265
G+ K + RE P D++ C C GH A +C + +C+ C K GH A
Sbjct: 509 GICHTCGKAGHRARECSAPPMPP-GDLRLCNNCYKQGHIAAECTNEKACNNCRKTGHLAR 685
Query: 266 ECKDVARDVTCYNCGEKGHISTKCTK 291
+C + D C C GH++ +C K
Sbjct: 686 DCPN---DPICNLCNVSGHVARQCPK 754
Score = 48.1 bits (113), Expect = 3e-05
Identities = 25/81 (30%), Positives = 30/81 (36%), Gaps = 23/81 (28%)
Frame = +2
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDV-----------------------A 271
C C GH A DC D C+ C +GH A +C
Sbjct: 650 CNNCRKTGHLARDCPNDPICNLCNVSGHVARQCPKANVLGDRSGGGGGGGGARGGGGGGY 829
Query: 272 RDVTCYNCGEKGHISTKCTKP 292
RDV C NC + GH+S C P
Sbjct: 830 RDVVCRNCQQLGHMSRDCMGP 892
Score = 40.4 bits (93), Expect = 0.006
Identities = 23/96 (23%), Positives = 36/96 (36%), Gaps = 26/96 (27%)
Frame = +2
Query: 220 REVGGGSKPSLADIKCYRCGTLGHYANDCKK--------------------------DIS 253
R+ G ++ D C C GH A C K D+
Sbjct: 662 RKTGHLARDCPNDPICNLCNVSGHVARQCPKANVLGDRSGGGGGGGGARGGGGGGYRDVV 841
Query: 254 CHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKC 289
C C + GH + +C + C+NCG +GH++ +C
Sbjct: 842 CRNCQQLGHMSRDCMGPL--MICHNCGGRGHLAYEC 943
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 59.3 bits (142), Expect = 1e-08
Identities = 28/82 (34%), Positives = 43/82 (52%), Gaps = 12/82 (14%)
Frame = +1
Query: 226 SKPSLADIKCYRCGTLGHYA-----NDCKKDISCHKCGKAGHKAAECKDVARDVT----- 275
S+ L +I+CY C LGH + +ISC+KCG+ GH C + ++T
Sbjct: 319 SQDDLKEIQCYVCKRLGHLCCVNTDDATPGEISCYKCGQLGHTGLACSRLRDEITSGATP 498
Query: 276 --CYNCGEKGHISTKCTKPKKA 295
C+ CGE+GH + +CT K+
Sbjct: 499 SSCFKCGEEGHFARECTSSIKS 564
Score = 51.6 bits (122), Expect = 2e-06
Identities = 23/60 (38%), Positives = 27/60 (44%), Gaps = 5/60 (8%)
Frame = +1
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARDVT-----CYNCGEKGHISTKC 289
CY CG LGH A C K C C K GH+A +C + + C CG GH C
Sbjct: 127 CYVCGCLGHNARQCSKVQDCFICKKGGHRAKDCPEKHTSTSKSIAICLKCGNSGHDIFSC 306
Score = 48.1 bits (113), Expect = 3e-05
Identities = 42/159 (26%), Positives = 63/159 (39%), Gaps = 17/159 (10%)
Frame = +1
Query: 235 CYRCGTLGHYANDC---KKDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCTK 291
C+ CG GH A +C K+ C+ CG GH A +C V C+ C + GH + C +
Sbjct: 61 CFNCGEEGHAAVNCSAVKRKKPCYVCGCLGHNARQCSKVQ---DCFICKKGGHRAKDCPE 231
Query: 292 PKKAAGKVFAL------------NAEEVEQPDNLIRGMCFI--NSTHLIGIIDIGATHSF 337
+ K A+ + D+L C++ HL + AT
Sbjct: 232 KHTSTSKSIAICLKCGNSGHDIFSCRNDYSQDDLKEIQCYVCKRLGHLCCVNTDDATPG- 408
Query: 338 ISVSCVKRLKLVVTPLLRGMVIDTPARGSVTTSFMCAKC 376
+SC K +L T L + D G+ +S C KC
Sbjct: 409 -EISCYKCGQLGHTGLACSRLRDEITSGATPSS--CFKC 516
Score = 43.5 bits (101), Expect = 7e-04
Identities = 17/39 (43%), Positives = 22/39 (55%)
Frame = +1
Query: 253 SCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCTK 291
+C CG+ GH A C V R CY CG GH + +C+K
Sbjct: 58 ACFNCGEEGHAAVNCSAVKRKKPCYVCGCLGHNARQCSK 174
Score = 36.2 bits (82), Expect = 0.11
Identities = 18/53 (33%), Positives = 28/53 (51%)
Frame = +1
Query: 217 RKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKD 269
R + E+ G+ PS C++CG GH+A +C I K GK +++ KD
Sbjct: 463 RLRDEITSGATPS----SCFKCGEEGHFARECTSSI---KSGKRNWESSHTKD 600
>TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24M7, partial (36%)
Length = 1224
Score = 57.8 bits (138), Expect = 3e-08
Identities = 36/104 (34%), Positives = 51/104 (48%), Gaps = 12/104 (11%)
Frame = +1
Query: 210 KPYNKDKRKKREV--GGGSKPSLADIKCYRCGTLGHYANDC------KKDISCHKCGKAG 261
+P K +KR + G KP + C+ C + H A C +K+ C +C + G
Sbjct: 199 RPEPKPGSRKRHLLRVPGMKPGES---CFICKAMDHIAKLCPEKAEWEKNKICLRCRRRG 369
Query: 262 HKAAECKDV---ARDVT-CYNCGEKGHISTKCTKPKKAAGKVFA 301
H+A C +V A+D CYNCGE GH T+C P + G FA
Sbjct: 370 HRAKNCPEVLDGAKDAKYCYNCGENGHALTQCLHPLQEGGTKFA 501
>TC233976
Length = 763
Score = 57.0 bits (136), Expect = 6e-08
Identities = 40/128 (31%), Positives = 53/128 (41%), Gaps = 16/128 (12%)
Frame = +1
Query: 183 KFDDAGRVKANYYKAQSEKRGKG-----HGVGKPYNKDKRKKREVGG----------GSK 227
+FD + K +YK + GK H KPY+ + GG G
Sbjct: 10 RFD*DQQEKVAFYKNANASHGKEKKPMTHSRAKPYSAPLEYENHYGGQRTSGGHHLAGGS 189
Query: 228 PSLADIKCYRCGTLGHYANDC-KKDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGHIS 286
L + G G A + C KCG+ GH A EC R+VTC+N KGH+S
Sbjct: 190 SQLVNRVSQPAGRGGSGAPAIVTTPLRCRKCGRLGHNAHEC--TYREVTCFNYQGKGHLS 363
Query: 287 TKCTKPKK 294
T C P+K
Sbjct: 364 TNCPHPRK 387
>TC227589
Length = 547
Score = 57.0 bits (136), Expect = 6e-08
Identities = 27/79 (34%), Positives = 39/79 (49%), Gaps = 12/79 (15%)
Frame = +2
Query: 226 SKPSLADIKCYRCGTLGHYA-----NDCKKDISCHKCGKAGHKAAECKDVARDVT----- 275
S L +I+CY C +GH + +ISC+KCG+ GH C + ++T
Sbjct: 305 SPDDLKEIQCYVCKRVGHLCCVNTDDATPGEISCYKCGQLGHTGLACSKLPDEITSAATP 484
Query: 276 --CYNCGEKGHISTKCTKP 292
C CGE GH + +CT P
Sbjct: 485 SSCCKCGEAGHFAQECTSP 541
Score = 53.9 bits (128), Expect = 5e-07
Identities = 45/149 (30%), Positives = 55/149 (36%), Gaps = 7/149 (4%)
Frame = +2
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAEC--KDVARD---VTCYNCGEKGHISTKC 289
CY CG LGH A C K C C K GH+A +C K +R C CG GH C
Sbjct: 113 CYVCGGLGHNARQCTKAQDCFICKKGGHRAKDCLEKHTSRSKSVAICLKCGNSGHDMFSC 292
Query: 290 TKPKKAAGKVFALNAEEVEQPDNLIRGMCFI--NSTHLIGIIDIGATHSFISVSCVKRLK 347
PD+L C++ HL + AT +SC K +
Sbjct: 293 RND---------------YSPDDLKEIQCYVCKRVGHLCCVNTDDATPG--EISCYKCGQ 421
Query: 348 LVVTPLLRGMVIDTPARGSVTTSFMCAKC 376
L T L + D S T C KC
Sbjct: 422 LGHTGLACSKLPDEIT--SAATPSSCCKC 502
Score = 43.9 bits (102), Expect = 5e-04
Identities = 17/39 (43%), Positives = 21/39 (53%)
Frame = +2
Query: 253 SCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCTK 291
+C CG+ GH A C R CY CG GH + +CTK
Sbjct: 44 ACFNCGEDGHAAVNCSAAKRKKPCYVCGGLGHNARQCTK 160
Score = 30.0 bits (66), Expect = 7.6
Identities = 9/19 (47%), Positives = 13/19 (68%)
Frame = +2
Query: 276 CYNCGEKGHISTKCTKPKK 294
C+NCGE GH + C+ K+
Sbjct: 47 CFNCGEDGHAAVNCSAAKR 103
>BU549069
Length = 615
Score = 53.1 bits (126), Expect = 8e-07
Identities = 40/128 (31%), Positives = 61/128 (47%)
Frame = -3
Query: 1282 ESESCNWSWSYFEV*EVDSSFCWAV*CC*DGWGCSVSDCVTIVFVESS*CVSCVSVEEIC 1341
ES S +W WS E+ + +S + * C + +C+T + SS C+SCVS +
Sbjct: 613 ESHSKDWGWSSTEIPKTHTSLYRSFPNS*KSXSCGIPNCITPITF*SSQCLSCVSTPYVY 434
Query: 1342 VRCVSCDSSGRIGSKR*FDR*DLAG*D*GP*SEAFAWQGDCFGQSGLGWTNW*ECYLGIG 1401
+SC G SKR D *++ D G ++A +G+ G LG C++GI
Sbjct: 433 P*SISCGQIG*RTSKRELDI*NITIEDRG*KNKAPKKEGESIG*GDLGRYIRRRCHVGIR 254
Query: 1402 E*DEGFVS 1409
E D +S
Sbjct: 253 ESDASSLS 230
>TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (42%)
Length = 655
Score = 49.3 bits (116), Expect = 1e-05
Identities = 25/78 (32%), Positives = 30/78 (38%), Gaps = 20/78 (25%)
Frame = +3
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDV--------------------ARDV 274
C C GH A DC D C+ C +GH A +C RDV
Sbjct: 135 CNNCRKTGHLARDCPNDPICNLCNVSGHVARQCPKANVLGDXXGGGGGARGGGGGGYRDV 314
Query: 275 TCYNCGEKGHISTKCTKP 292
C NC + GH+S C P
Sbjct: 315 VCRNCQQLGHMSRDCMGP 368
Score = 48.5 bits (114), Expect = 2e-05
Identities = 20/57 (35%), Positives = 29/57 (50%)
Frame = +3
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCTK 291
C C GH A +C + +C+ C K GH A +C + D C C GH++ +C K
Sbjct: 78 CNNCYKQGHIAAECTNEKACNNCRKTGHLARDCPN---DPICNLCNVSGHVARQCPK 239
Score = 43.5 bits (101), Expect = 7e-04
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 7/61 (11%)
Frame = +3
Query: 236 YRCGTLGHYANDCKK------DIS-CHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTK 288
+ CG GH A +C D+ C+ C K GH AAEC + C NC + GH++
Sbjct: 3 HTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC---TNEKACNNCRKTGHLARD 173
Query: 289 C 289
C
Sbjct: 174 C 176
Score = 42.4 bits (98), Expect = 0.001
Identities = 19/43 (44%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Frame = +3
Query: 255 HKCGKAGHKAAECKDVARDV----TCYNCGEKGHISTKCTKPK 293
H CGKAGH+A EC C NC ++GHI+ +CT K
Sbjct: 3 HTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAECTNEK 131
Score = 41.6 bits (96), Expect = 0.003
Identities = 23/93 (24%), Positives = 36/93 (37%), Gaps = 23/93 (24%)
Frame = +3
Query: 220 REVGGGSKPSLADIKCYRCGTLGHYANDCKK-----------------------DISCHK 256
R+ G ++ D C C GH A C K D+ C
Sbjct: 147 RKTGHLARDCPNDPICNLCNVSGHVARQCPKANVLGDXXGGGGGARGGGGGGYRDVVCRN 326
Query: 257 CGKAGHKAAECKDVARDVTCYNCGEKGHISTKC 289
C + GH + +C + C+NCG +GH++ +C
Sbjct: 327 CQQLGHMSRDCMGPL--MICHNCGGRGHLAYEC 419
>CF922531
Length = 602
Score = 46.6 bits (109), Expect = 8e-05
Identities = 43/136 (31%), Positives = 66/136 (47%), Gaps = 13/136 (9%)
Frame = -2
Query: 423 SKPVEKLDRKFLTAEQVKKSLDGEACVFMMFASLKENSEKGVGDL----PIVQE------ 472
SK + K + F T +K +L + +++ + S + + P VQE
Sbjct: 409 SKKIIKKENHFATKGDIKIALLLKQSFYLLLSRETSLSTVTIPTIETLPPKVQELLHEFG 230
Query: 473 --FPEDIT-ELPLEREVEFAIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRP 529
FP++I LP R +E IDLVP S Y + E E++ Q++ELLEK +++
Sbjct: 229 DIFPKEIPLGLPPLRGIEHQIDLVPRASLPNRPTYRTNPQETKEIESQVKELLEKGWVQE 50
Query: 530 SVSPWGAPVLLVKKKD 545
S+S VLLV KKD
Sbjct: 49 SLSLCVVLVLLVPKKD 2
>TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequence, partial
(22%)
Length = 742
Score = 46.2 bits (108), Expect = 1e-04
Identities = 29/100 (29%), Positives = 40/100 (40%), Gaps = 21/100 (21%)
Frame = +2
Query: 219 KREVGGGSKPSLADIKCYRCGTLGHYANDCKKDIS-----------CHKCGKAGHKAAEC 267
+R GGG+ CY+CG GH A DC + + C CG GH A +C
Sbjct: 17 RRNGGGGAA-------CYQCGEFGHLARDCNRSSNSGGGGGGSGGGCFNCGGFGHLARDC 175
Query: 268 -KDVARDV---------TCYNCGEKGHISTKCTKPKKAAG 297
+ V +C+ CG GH++ C K G
Sbjct: 176 VRGGGGSVGIGGGGGGGSCFRCGGFGHMARDCATGKGNIG 295
Score = 44.3 bits (103), Expect = 4e-04
Identities = 32/102 (31%), Positives = 41/102 (39%), Gaps = 16/102 (15%)
Frame = +2
Query: 219 KREVGGGSKPSLADIKCYRCGTLGHYANDCKKD----------------ISCHKCGKAGH 262
K +GGG C+RCG +GH A DC + +C CGK GH
Sbjct: 281 KGNIGGGGSGG----GCFRCGEVGHLARDCGMEGGRFGGGGGSGGGGGKSTCFNCGKPGH 448
Query: 263 KAAECKDVARDVTCYNCGEKGHISTKCTKPKKAAGKVFALNA 304
A EC V GE+G + T C KK +F+ A
Sbjct: 449 FAREC------VEASG*GEEGFV-TFCEGRKKVE*YIFSFLA 553
>CD487724
Length = 676
Score = 46.2 bits (108), Expect = 1e-04
Identities = 52/221 (23%), Positives = 93/221 (41%), Gaps = 22/221 (9%)
Frame = +3
Query: 325 LIGIIDIGATHSFISVSCVKRLKLVV--TPLLRGMVIDTPARGSVTTSFMCAKCPVNFGN 382
L+ ++D G+TH+F+ V +L L TP LR MV + T +C P++ N
Sbjct: 24 LVYLVDGGSTHNFVQQPLVSQLGLPCRSTPPLRVMVGNGHHLKCTT---ICEAIPISIQN 194
Query: 383 VDFELDLVCLPLKHMDVIFGMDWLLFFG---VSINCLTKSVTFSKPVEKLDRK------F 433
++F + L LP+ +++ G+ WL G V N L+ + + +L +
Sbjct: 195 IEFLVHLYVLPIVGANIVLGVQWLKTLGPILVDYNSLSMQFFYQHRLVQLKGESEAQLGL 374
Query: 434 LTAEQVKK-SLDGEACVFMMFASLKENSEKGVGDLPIVQEFPEDITE----------LPL 482
L Q+++ E + A L EN+ P+ Q + + LP
Sbjct: 375 LNHHQLRRLHQTHEPVTYFHIAILTENTSP-TSSPPLPQPIQHLLDQFSALFQ*PQGLPP 551
Query: 483 EREVEFAIDLVPGMSPIWITPYPMSASELGELKKQLEELLE 523
RE + I L+P P+ + Y E++ Q+ +L+
Sbjct: 552 ARETDHHIHLLP*SEPVNMRLY*YPHY*NNEIEHQVNLMLQ 674
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo}, partial
(9%)
Length = 410
Score = 45.8 bits (107), Expect = 1e-04
Identities = 42/136 (30%), Positives = 62/136 (44%), Gaps = 8/136 (5%)
Frame = -2
Query: 1262 ELS**EEKGY*VSGR*SCVFESESCNWSWSYFEV*EVDSSFCWAV*CC*DGWGCSVSDCV 1321
ELS EE+ + G *SC+ +S +W WS FE+ + +SF + C +S+C+
Sbjct: 409 ELSRQEEERPRIRGW*SCILKSHPIDWGWSSFEILKTHTSFYRSFPNSQKS*FCGISNCI 230
Query: 1322 TIVFVESS*CVSCVSVEEIC--------VRCVSCDSSGRIGSKR*FDR*DLAG*D*GP*S 1373
T + S C+SC S I RC C S G G+ *++A D G +
Sbjct: 229 TPISY*PSQCLSCFSNP*IY**SFSHGQFRC--CSSKGEFGT------*NIALEDQGYAN 74
Query: 1374 EAFAWQGDCFGQSGLG 1389
+A + D G LG
Sbjct: 73 KALKRERDFVG*GNLG 26
>TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like protein, partial
(62%)
Length = 1131
Score = 44.7 bits (104), Expect = 3e-04
Identities = 26/82 (31%), Positives = 34/82 (40%), Gaps = 1/82 (1%)
Frame = +1
Query: 214 KDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARD 273
+ R + +G G P +C+ CG GH+A DCK +KC
Sbjct: 307 RGSRDREYMGRGPPPGSG--RCFNCGIDGHWARDCKAGDWKNKC---------------- 432
Query: 274 VTCYNCGEKGHISTKC-TKPKK 294
Y CGE+GHI C PKK
Sbjct: 433 ---YRCGERGHIEKNCKNSPKK 489
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.350 0.157 0.564
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,791,938
Number of Sequences: 63676
Number of extensions: 1085626
Number of successful extensions: 9775
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 6874
Number of HSP's successfully gapped in prelim test: 245
Number of HSP's that attempted gapping in prelim test: 2409
Number of HSP's gapped (non-prelim): 7653
length of query: 1419
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1310
effective length of database: 5,698,948
effective search space: 7465621880
effective search space used: 7465621880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC144724.7


