

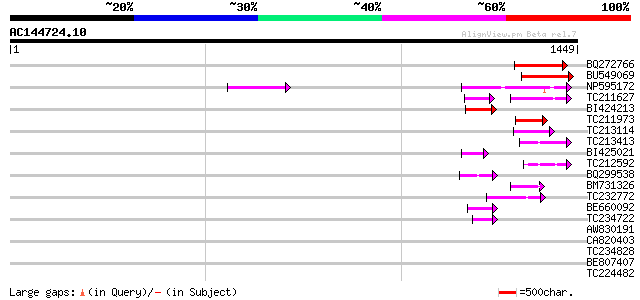
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144724.10 - phase: 0 /pseudo
(1449 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 193 4e-49
BU549069 186 8e-47
NP595172 polyprotein [Glycine max] 158 2e-38
TC211627 86 5e-32
BI424213 78 2e-14
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 75 3e-13
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 70 7e-12
TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 67 4e-11
BI425021 61 4e-09
TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 59 2e-08
BQ299538 58 3e-08
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 55 2e-07
TC232772 weakly similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial ... 52 2e-06
BE660092 weakly similar to GP|9884624|dbj retroelement pol polyp... 51 4e-06
TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (... 46 1e-04
AW830191 43 0.001
CA820403 weakly similar to GP|13273463|gb| pol protein integrase... 39 0.017
TC234828 37 0.048
BE807407 34 0.41
TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 33 0.91
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo}, partial
(9%)
Length = 410
Score = 193 bits (491), Expect = 4e-49
Identities = 93/136 (68%), Positives = 112/136 (81%)
Frame = -3
Query: 1289 SYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQISERVGTVAYRVGL 1348
SYHDKRRKDLEF+ GDHVFLRVTP TGVGRALKS KLTP FIGP+QI ++V VAY++ L
Sbjct: 408 SYHDKRRKDLEFEVGDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIAL 229
Query: 1349 PPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLRIDDRKVKTLRGKEI 1408
PP L++LH+VFHVSQ+ KY+ DPSH++ D VQV++NL ETLPLRI D + K LRGKEI
Sbjct: 228 PPSLTSLHNVFHVSQIHKYINDPSHMVNLDVVQVKENLAHETLPLRIKDMRTKHLRGKEI 49
Query: 1409 PLVRVVWDGATGESLT 1424
LV+V+W GA+GE T
Sbjct: 48 LLVKVIWGGASGEEAT 1
>BU549069
Length = 615
Score = 186 bits (471), Expect = 8e-47
Identities = 88/132 (66%), Positives = 108/132 (81%)
Frame = -1
Query: 1308 LRVTPVTGVGRALKSKKLTPRFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKY 1367
L+VTP TGVG+ALKS+KLTP FIG +QI +R VAY++ LPP LSNLH+VFHVSQLR Y
Sbjct: 615 LKVTPRTGVGQALKSRKLTPHFIGHFQILKRAXPVAYQIALPPSLSNLHNVFHVSQLRMY 436
Query: 1368 VPDPSHVIQSDDVQVRDNLTVETLPLRIDDRKVKTLRGKEIPLVRVVWDGATGESLTWEL 1427
+ DPSHV++ DDVQV++NLT ETLPLRI+DR+ K LR KE PLV+V+W G +GE TWEL
Sbjct: 435 IHDPSHVVKLDDVQVKENLTYETLPLRIEDRRTKHLRRKENPLVKVIWGGTSGEDATWEL 256
Query: 1428 ESKMLESYPKLF 1439
ES+M +YP LF
Sbjct: 255 ESQMRVAYPSLF 220
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 158 bits (399), Expect = 2e-38
Identities = 103/297 (34%), Positives = 152/297 (50%), Gaps = 15/297 (5%)
Frame = +1
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRD FTS FW+ L + G+ L +SSAYHPQ+DGQSE + LE LR E W
Sbjct: 3619 VSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCFTYEHPKGW 3798
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRT---QLCWFESGGRVVLGPEIVQQTTE 1271
LP EF YN +YH S+GM PF ALYGR T Q C + E+ +Q T+
Sbjct: 3799 VKALPWAEFWYNTAYHMSLGMTPFRALYGREPPTLTRQACSIDDPA------EVREQLTD 3960
Query: 1272 KVKMIQE---KMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRAL-KSKKLTP 1327
+ ++ + + +Q K DK+R D+ FQ GD V +++ P L K++KL+
Sbjct: 3961 RDALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHSAVLRKNQKLSM 4140
Query: 1328 RFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLR--------KYVPDPSHVIQSDD 1379
R+ GP+++ ++G VAY++ L P + +H VFHVSQL+ Y+P P V +
Sbjct: 4141 RYFGPFKVLAKIGDVAYKLEL-PSAARIHPVFHVSQLKPFNGTAQDPYLPLPLTVTEMGP 4317
Query: 1380 VQVRDNLTVETLPLRIDDRKVKTLRGKEIPLVRVVWDGATGESLTWELESKMLESYP 1436
V P++I ++ +I + V W+ + TWE + SYP
Sbjct: 4318 VM---------QPVKILASRIIIRGHNQIEQILVQWENGLQDEATWEDIEDIKASYP 4461
Score = 45.8 bits (107), Expect = 1e-04
Identities = 42/161 (26%), Positives = 75/161 (46%)
Frame = +3
Query: 557 EERW*YEVVYRLSSVEQSNYQE*VSTSEN**FDGSVSGCTHFQQD*FEIRLSPD*SKR*R 616
EERW E+++RL S E ++ + *+S + + * + + F + FE+R+ + *
Sbjct: 1923 EERWKLEILHRL*SFECNHCEG*LSNANSG*TAR*TTWSSIFFKTGFEVRIPSNSGTA*G 2102
Query: 617 YAEDSFQNAVWSL*I*SYAFRCYECTWSVYGVHESHLSCILGSIRGCFYL*YFNLLQD*R 676
E+ F N W L*+ S A Y CT + ++E ++S I C + *+ ++
Sbjct: 2103 *RENCF*NTSWPL*MVSDAIWSY*CTSHIPMLNEQNISVCSQKICSCVF**HLDI*CFLE 2282
Query: 677 RTC*ASEDCLASVKREETLC*IIQV*VLVERGKFSWPCYFW 717
+ A C ++ +C*I+Q+ + R P W
Sbjct: 2283 GSSQALGICATDLEATSVIC*IVQMFIWRYRS*LLGPQGIW 2405
>TC211627
Length = 1034
Score = 86.3 bits (212), Expect(2) = 5e-32
Identities = 55/158 (34%), Positives = 79/158 (49%), Gaps = 1/158 (0%)
Frame = +3
Query: 1279 KMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQISER 1338
+++ Q K D R+DL F GD V++R+ P KL+ RF GPYQI R
Sbjct: 357 RLQKYQDSMKRIADSHRRDLTFNIGDWVYVRL*PYRQTSIQSTYTKLSKRFYGPYQIQAR 536
Query: 1339 VGTVAYRVGLPPHLSNLHDVFHVSQLR-KYVPDPSHVIQSDDVQVRDNLTVETLPLRIDD 1397
VG VAYR+ LPP S +H +FHVS L+ + P P ++ ++ V+ PL+ D
Sbjct: 537 VGQVAYRLQLPP-TSKIHPIFHVSLLKVHHGPIPPELLALPPFSTTNHPLVQ--PLQFLD 707
Query: 1398 RKVKTLRGKEIPLVRVVWDGATGESLTWELESKMLESY 1435
K+ IP V V W E TWE +++ + Y
Sbjct: 708 WKMDESTTPPIPQVLVQWTNLAPEDTTWESWTQLKDIY 821
Score = 71.6 bits (174), Expect(2) = 5e-32
Identities = 38/78 (48%), Positives = 44/78 (55%)
Frame = +2
Query: 1162 FTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLI 1221
F S W L G+KLR S+AYHPQTDGQ+E + LE LR V + W L L
Sbjct: 5 FISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSLA 184
Query: 1222 EFTYNNSYHSSIGMAPFE 1239
E YN S HS IG +PFE
Sbjct: 185 E*CYNTSVHSGIGFSPFE 238
>BI424213
Length = 426
Score = 78.2 bits (191), Expect = 2e-14
Identities = 36/80 (45%), Positives = 49/80 (61%)
Frame = +1
Query: 1164 SRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEF 1223
S FWK+L LG+KL S+ HPQTDGQ++ +SL LLR + +WD +LP +EF
Sbjct: 4 SHFWKTLWAKLGTKLLFSTTCHPQTDGQTKVVNRSLSTLLRALLKGNHKSWDEYLPHVEF 183
Query: 1224 TYNNSYHSSIGMAPFEALYG 1243
YN H + +PFE +YG
Sbjct: 184 AYNRGVHRTTKQSPFEVVYG 243
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 74.7 bits (182), Expect = 3e-13
Identities = 37/84 (44%), Positives = 58/84 (69%), Gaps = 1/84 (1%)
Frame = +1
Query: 1292 DKRRKDLEFQEGDHVFLRVTPVTGVGRALK-SKKLTPRFIGPYQISERVGTVAYRVGLPP 1350
+KRR+D+E+ GD VFL++ P A + ++KL+PRF P+Q+ +VGT+AY++ LP
Sbjct: 112 NKRRRDIEYVVGD*VFLKMQPYRRRSLAKRINEKLSPRFYAPFQVFNKVGTIAYKLDLPS 291
Query: 1351 HLSNLHDVFHVSQLRKYVPDPSHV 1374
H+ +H VFHVS L+K P+ H+
Sbjct: 292 HI-KIHPVFHVSLLKKASPNLYHL 360
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 70.1 bits (170), Expect = 7e-12
Identities = 42/115 (36%), Positives = 67/115 (57%), Gaps = 11/115 (9%)
Frame = +3
Query: 1288 KSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALK-SKKLTPRFIGPYQISERVGTVAYRV 1346
K+Y D+ R+ + GD V+L++ P A K ++KL+PRF GPYQI +++G VA+ +
Sbjct: 6 KAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFEL 185
Query: 1347 GLPPHLSNLHDVFHVSQLRKYV-----PDPSHVIQSDDV-----QVRDNLTVETL 1391
LPP +H VFH S L+K V P P ++ S+D+ Q++ L++ L
Sbjct: 186 DLPP-ARKIHPVFHASLLKKAVAATANPQPLPLMLSEDLSSEFFQLKSKLSITIL 347
>TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (5%)
Length = 761
Score = 67.4 bits (163), Expect = 4e-11
Identities = 43/134 (32%), Positives = 64/134 (47%), Gaps = 1/134 (0%)
Frame = +1
Query: 1303 GDHVFLRVTPVT-GVGRALKSKKLTPRFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHV 1361
GD V +++ P G KLT R+ GP+++ ER+G V YR+ L H S +H VFHV
Sbjct: 7 GDWVLVKLRPHRQGSASETTYSKLTKRYYGPFEVQERLGKVVYRLKLTAH-SRIHPVFHV 183
Query: 1362 SQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLRIDDRKVKTLRGKEIPLVRVVWDGATGE 1421
S L+ +V DP + + V PL + D K+ +V V W A+ +
Sbjct: 184 SLLKAFVGDP-ETTHAGPLPVMRTEEATNTPLTVIDSKLVPADNGPRRMVLVQWPSASRQ 360
Query: 1422 SLTWELESKMLESY 1435
+WE + E Y
Sbjct: 361 DASWEDWQVLRERY 402
>BI425021
Length = 426
Score = 60.8 bits (146), Expect = 4e-09
Identities = 31/68 (45%), Positives = 41/68 (59%)
Frame = -1
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRDP F S FW+ L G+ LR+SSAYHPQTDGQ+E + +E LR V +
Sbjct: 204 VSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVIEQYLRAFVHGRPRNL 25
Query: 1215 DSHLPLIE 1222
+P +E
Sbjct: 24 GRFIPWVE 1
>TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 664
Score = 58.5 bits (140), Expect = 2e-08
Identities = 42/124 (33%), Positives = 64/124 (50%)
Frame = +2
Query: 1312 PVTGVGRALKSKKLTPRFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDP 1371
P+TG KL+ RF GP+Q+ ERVG VAYR+ LP + LH VFH S L+ + +P
Sbjct: 62 PITG--------KLSKRFFGPFQVVERVGKVAYRLQLPVD-AKLHPVFHCSLLKPFQGNP 214
Query: 1372 SHVIQSDDVQVRDNLTVETLPLRIDDRKVKTLRGKEIPLVRVVWDGATGESLTWELESKM 1431
+ D+ +V PL I +T+ +I V V W G + + TWE +++
Sbjct: 215 PDTAAPLPPTLFDHQSV-IAPLVI--LATRTVNDDDIE-VLVQWQGLSPDDATWEKWTEL 382
Query: 1432 LESY 1435
+ +
Sbjct: 383 CKEF 394
>BQ299538
Length = 426
Score = 57.8 bits (138), Expect = 3e-08
Identities = 33/97 (34%), Positives = 49/97 (50%)
Frame = +3
Query: 1149 WCSFEHLSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVL 1208
WCS R F + + + G+ L++S++YHP DGQ+ LE LR V
Sbjct: 66 WCSGFSPQ*RGSNFCEFILEGIVKLPGTYLKMSTSYHP*IDGQTVN--HCLETFLRCFVA 239
Query: 1209 EQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRR 1245
+Q L E+ YN ++H+S G PFE +YGR+
Sbjct: 240 DQPKM*VQWLSWAEYWYNTNFHASTGTTPFEVVYGRK 350
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 55.5 bits (132), Expect = 2e-07
Identities = 31/88 (35%), Positives = 50/88 (56%), Gaps = 1/88 (1%)
Frame = +1
Query: 1280 MKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVT-GVGRALKSKKLTPRFIGPYQISER 1338
++ +QS + + R+ + GD V+L++ P G KLT RF GPY + +
Sbjct: 25 LERAQSLMVKHANNHRRPHDINVGDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQ 204
Query: 1339 VGTVAYRVGLPPHLSNLHDVFHVSQLRK 1366
VG VA+++ LP + +H VFHVSQL++
Sbjct: 205 VGAVAFQLQLPSE-ARIHPVFHVSQLKR 285
>TC232772 weakly similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (7%)
Length = 729
Score = 52.0 bits (123), Expect = 2e-06
Identities = 38/155 (24%), Positives = 69/155 (44%), Gaps = 4/155 (2%)
Frame = +1
Query: 1218 LPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVKMIQ 1277
LP +EF YN + HS+ PFE +Y T L + E +K +
Sbjct: 7 LPHVEFAYNRAVHSTTQHFPFEVVYDFNPLTPLDSLPLSNISGFKHKDAHAKVEYIKRLH 186
Query: 1278 EKMKASQSRQKSYH----DKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPY 1333
E+ K +++ + +K RK + + D V++ + + + KL PR GP+
Sbjct: 187 EQAKTQIAKKNESYVKQTNKNRKKVVLEPSDWVWVHMRKERFPKQRMS--KLQPRGDGPF 360
Query: 1334 QISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYV 1368
Q+ ER+ AY++ +P + F+++ L +V
Sbjct: 361 QVLERINYNAYKIDIPGEY-EVSSSFNIADLTPFV 462
>BE660092 weakly similar to GP|9884624|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (13%)
Length = 378
Score = 50.8 bits (120), Expect = 4e-06
Identities = 23/78 (29%), Positives = 44/78 (55%)
Frame = -3
Query: 1169 SLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNS 1228
+L + G R+S+ YHPQT+GQ+E + + ++ +L V W + L + + +
Sbjct: 370 ALLKKYGVVHRVSTPYHPQTNGQAEISNREIKRILEKIVQPSRKDWSTRLDDALWAHRTA 191
Query: 1229 YHSSIGMAPFEALYGRRC 1246
Y + IGM+P+ ++G+ C
Sbjct: 190 YKAPIGMSPYRVVFGKAC 137
>TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (32%)
Length = 482
Score = 46.2 bits (108), Expect = 1e-04
Identities = 20/63 (31%), Positives = 37/63 (57%)
Frame = -3
Query: 1184 YHPQTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYG 1243
YHPQT+GQ+E + + ++ +L V+ W L + Y ++ + IG++PF+ +YG
Sbjct: 234 YHPQTNGQAEVSNKEIKRVLENIVVSSRKDWALKLDDAFWAYRIAFKTPIGLSPFQLVYG 55
Query: 1244 RRC 1246
+ C
Sbjct: 54 KAC 46
>AW830191
Length = 372
Score = 42.7 bits (99), Expect = 0.001
Identities = 27/103 (26%), Positives = 48/103 (46%), Gaps = 4/103 (3%)
Frame = +3
Query: 1214 WDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVV--LGPEIVQQTTE 1271
W L EF +N +Y++S+ + PF+ LYG L G ++ E+ T +
Sbjct: 39 WPKRLSWAEFWFNTNYNNSLKLTPFKVLYGCDPPHLL-----KGAIISSTAEEVNVMTND 203
Query: 1272 KVKMIQEKM--KASQSRQKSYHDKRRKDLEFQEGDHVFLRVTP 1312
+ +M+ + A Q Y D+ R+ + GD V+L++ P
Sbjct: 204 RDQMLHDLKGNLAKAQNQMKYADRSRRSIPLNVGDWVYLKLQP 332
>CA820403 weakly similar to GP|13273463|gb| pol protein integrase region
{Ginkgo biloba}, partial (52%)
Length = 421
Score = 38.9 bits (89), Expect = 0.017
Identities = 24/70 (34%), Positives = 35/70 (49%), Gaps = 1/70 (1%)
Frame = -3
Query: 1146 KVAWCSFEHLSDRDPRFT-SRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLR 1204
K+ CS +SD D F S FW L + G+KL+ S AYHPQ D ++ + +E L+
Sbjct: 317 KLHGCSSSIVSDWDRLFLIS*FWTELFKMEGTKLKFSLAYHPQPDSHTKVVNRCIEMNLQ 138
Query: 1205 VCVLEQGGTW 1214
+ W
Sbjct: 137 CLTTSKRKQW 108
>TC234828
Length = 857
Score = 37.4 bits (85), Expect = 0.048
Identities = 30/74 (40%), Positives = 39/74 (52%)
Frame = +2
Query: 478 TSGVRFS*SFSR*NS*CASRERS*VFY*SYSWNGAGVDGTLSYISFRVS*VEETVRRFA* 537
T GVR S SFS * * A+ ERS + Y* +W + VD L +S S E T
Sbjct: 632 TGGVRISRSFSG*CL*IAT*ERSGIHY*RGAWGESSVDCAL*NVSGGTSRGEGTSTGPFE 811
Query: 538 EEVC*TKCFTLGSA 551
+ +C +K T+GSA
Sbjct: 812 QTICSSKRITVGSA 853
>BE807407
Length = 341
Score = 34.3 bits (77), Expect = 0.41
Identities = 14/28 (50%), Positives = 20/28 (71%)
Frame = +2
Query: 1324 KLTPRFIGPYQISERVGTVAYRVGLPPH 1351
KLT R+ GPY + +VG VA+++ LP H
Sbjct: 158 KLTARYYGPYLVLIKVGVVAFQLQLPKH 241
>TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 669
Score = 33.1 bits (74), Expect = 0.91
Identities = 34/137 (24%), Positives = 52/137 (37%), Gaps = 15/137 (10%)
Frame = +1
Query: 1214 WDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRC--------RTQLCWFESGGRVVLGPEI 1265
W LP Y S +S G PF +YG + ESG + E
Sbjct: 61 WHEMLPFALHGYRTSVRTSTGATPFSLVYGMEAVLPFEVEVPSLRILAESG---LKESEW 231
Query: 1266 VQQTTEKVKMIQEKM-------KASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGR 1318
Q +++ +I+ K + Q R KS DK+ +F EGD V +++ R
Sbjct: 232 AQTRYDQLNLIEGKRLTAMSHGRLYQQRMKSAFDKKVCLRKFHEGDLVLKKMSHAVKDHR 411
Query: 1319 ALKSKKLTPRFIGPYQI 1335
K P + GP+ +
Sbjct: 412 G----KWAPNYEGPFVV 450
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.362 0.162 0.603
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 67,530,328
Number of Sequences: 63676
Number of extensions: 1008402
Number of successful extensions: 10287
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 5545
Number of HSP's successfully gapped in prelim test: 335
Number of HSP's that attempted gapping in prelim test: 4481
Number of HSP's gapped (non-prelim): 6318
length of query: 1449
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1340
effective length of database: 5,698,948
effective search space: 7636590320
effective search space used: 7636590320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (22.0 bits)
S2: 65 (29.6 bits)
Medicago: description of AC144724.10


