

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.9 + phase: 0
(259 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
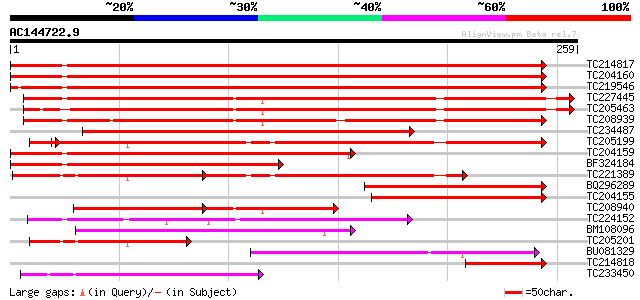
Score E
Sequences producing significant alignments: (bits) Value
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 358 1e-99
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 357 3e-99
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 347 2e-96
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 295 1e-80
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 283 7e-77
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 270 6e-73
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 247 3e-66
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 207 8e-56
TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 203 7e-53
BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chapero... 172 1e-43
TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa p... 100 1e-38
BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cyto... 134 3e-32
TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, ... 131 3e-31
TC208940 UP|Q39803 (Q39803) BiP isoform C (Fragment), partial (26%) 91 4e-27
TC224152 similar to UP|Q8L8N8 (Q8L8N8) 70kD heat shock protein, ... 98 4e-21
BM108096 95 4e-20
TC205201 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa prote... 74 7e-14
BU081329 70 1e-12
TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous hea... 70 1e-12
TC233450 65 2e-11
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 358 bits (920), Expect = 1e-99
Identities = 179/245 (73%), Positives = 205/245 (83%)
Frame = +2
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+V FTD +RL+G AA
Sbjct: 86 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 259
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ DI WPFKVI G DKPMI V YKG+EK
Sbjct: 260 KNQVAMNPINTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIPGAADKPMIVVNYKGEEKQF 439
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEAYL S VKNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN
Sbjct: 440 AAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 619
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 620 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 799
Query: 241 FDNRM 245
FDNRM
Sbjct: 800 FDNRM 814
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 357 bits (916), Expect = 3e-99
Identities = 177/245 (72%), Positives = 206/245 (83%)
Frame = +2
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 95 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 268
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ D+ WPFKVI G +KPMI V YKG+EK
Sbjct: 269 KNQVAMNPTNTVFDAKRLIGRRFSDASVQGDMKLWPFKVIPGPAEKPMIVVNYKGEEKQF 448
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEAYL S +KNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 449 SAEEISSMVLMKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 628
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 629 EPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 808
Query: 241 FDNRM 245
FDNRM
Sbjct: 809 FDNRM 823
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 347 bits (891), Expect = 2e-96
Identities = 173/245 (70%), Positives = 205/245 (83%)
Frame = +1
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MATK-EGKAIGIDLGTTYSCVGVW--QNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAA 171
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NPQNTVFDAKRLIGR+FSD VQ D+ WPFKV DKPMI V YKG+EK
Sbjct: 172 KNQVAMNPQNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCDKPMIVVNYKGEEKKF 351
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M+++AEA+L VKNAVVTVPAYFNDSQR+AT +AGAI+GLNV+RIIN
Sbjct: 352 SAEEISSMVLVKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIIN 531
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVS+LTI+ +FEVKATAG+THLGGED
Sbjct: 532 EPTAAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGED 711
Query: 241 FDNRM 245
FDNRM
Sbjct: 712 FDNRM 726
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 295 bits (755), Expect = 1e-80
Identities = 158/253 (62%), Positives = 194/253 (76%), Gaps = 1/253 (0%)
Frame = +2
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDLGTTYSCVGV+ + VEII ND+GN+ITPS+VAFTD +RL+G AAK+ AA+
Sbjct: 209 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVAFTDSERLIGEAAKNLAAV 382
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYK-GQEKNLCAEEI 125
NP+ T+FD KRLIGRKF D VQ+D+ P+K+++ + KP I VK K G+ K EEI
Sbjct: 383 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNK-DGKPYIQVKIKDGETKVFSPEEI 559
Query: 126 SSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAA 185
S++IL MK+ AEA+L + +AVVTVPAYFND+QR+AT +AG IAGLNV RIIN+PTAA
Sbjct: 560 SAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 739
Query: 186 AIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
AIAYGLD KG GE+NI VFDLGGGTFDVS+LTI VFEV AT G+THLGGEDFD R+
Sbjct: 740 AIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 910
Query: 246 CWNKMCFTMNLIK 258
M + + LIK
Sbjct: 911 ----MEYFIKLIK 937
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 283 bits (723), Expect = 7e-77
Identities = 157/253 (62%), Positives = 190/253 (75%), Gaps = 1/253 (0%)
Frame = +3
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDL TTYSCVGV + VEII ND+GN+ITPS+VAFTD +RL+G AAK AA+
Sbjct: 192 GTVIGIDL-TTYSCVGV----TDAVEIIANDQGNRITPSWVAFTDSERLIGEAAKIVAAV 356
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYK-GQEKNLCAEEI 125
NP T+FD KRLIGRKF D VQ+D+ P+K+++ + KP I VK K G+ K EEI
Sbjct: 357 NPVRTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNK-DGKPYIQVKIKDGETKVFSPEEI 533
Query: 126 SSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAA 185
S++IL MK+ AEA+L + +AVVTVPAYFND+QR+AT +AG IAGLNV RIIN+PTAA
Sbjct: 534 SAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 713
Query: 186 AIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
AIAYGLD KG GE+NI VFDLGGGTFDVS+LTI VFEV AT G+THLGGEDFD R+
Sbjct: 714 AIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 884
Query: 246 CWNKMCFTMNLIK 258
M + + LIK
Sbjct: 885 ----MEYFIKLIK 911
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 270 bits (689), Expect = 6e-73
Identities = 146/240 (60%), Positives = 181/240 (74%), Gaps = 1/240 (0%)
Frame = +3
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDLGTTYSCVGV+ + VEII ND+GN+ITPS+ +FTD +RL+G AAK+ AA+
Sbjct: 171 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSW-SFTDSERLIGEAAKNLAAV 341
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYK-GQEKNLCAEEI 125
NP+ +FD KRLIGRKF D VQ+D+ P+K+++ + KP I K K G+ K EEI
Sbjct: 342 NPERVIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNK-DGKPYIQEKIKDGETKVFSPEEI 518
Query: 126 SSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAA 185
S++IL MK+ AEA+L + +AV AYFND+QR+AT +AG IAGLNV RIIN+PTAA
Sbjct: 519 SAMILTKMKETAEAFLGKKINDAV----AYFNDAQRQATKDAGVIAGLNVARIINEPTAA 686
Query: 186 AIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
AIAYGLD KG GE+NI VFDLGGGTFDVS+LTI VFEV AT G+THLGGEDFD R+
Sbjct: 687 AIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 857
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 247 bits (631), Expect = 3e-66
Identities = 118/152 (77%), Positives = 140/152 (91%)
Frame = +1
Query: 34 IHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIM 93
IHND+GN TPS VAFTD QRL+G AAK+QAA NP+NTVFDAKRLIGRKFSDPV+QKD M
Sbjct: 1 IHNDQGNNTTPSCVAFTDQQRLIGEAAKNQAATNPENTVFDAKRLIGRKFSDPVIQKDKM 180
Query: 94 FWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSIILANMKKIAEAYLKSPVKNAVVTVP 153
WPFKV++G+NDKPMI++ YKGQEK+L AEE+SS++L M++IAEAYL++PV+NAVVTVP
Sbjct: 181 LWPFKVVAGINDKPMISLNYKGQEKHLLAEEVSSMVLIKMREIAEAYLETPVENAVVTVP 360
Query: 154 AYFNDSQRKATMNAGAIAGLNVMRIINDPTAA 185
AYFNDSQRKAT++AGAIAGLNVMRIIN+PTAA
Sbjct: 361 AYFNDSQRKATIDAGAIAGLNVMRIINEPTAA 456
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 207 bits (528), Expect(2) = 8e-56
Identities = 115/227 (50%), Positives = 149/227 (64%), Gaps = 1/227 (0%)
Frame = +3
Query: 20 CVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDD-QRLVGGAAKDQAAINPQNTVFDAKRL 78
C +L ++I N EG + TPS VAF + LVG AK QA NP NT+F KRL
Sbjct: 150 CAD*YLLHFQNPKVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTKRL 329
Query: 79 IGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSIILANMKKIAE 138
IGR+F D QK++ P+K++ N V+ GQ+ + ++ + +L MK+ AE
Sbjct: 330 IGRRFDDSQTQKEMKMVPYKIVKAPNGDAW--VEANGQQYS--PSQVGAFVLTKMKETAE 497
Query: 139 AYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIAYGLDNKGRCD 198
+YL V AV+TVPAYFND+QR+AT +AG IAGL+V RIIN+PTAAA++YG++NK
Sbjct: 498 SYLGKSVSKAVITVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNNK---- 665
Query: 199 GERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
E I VFDLGGGTFDVS+L I VFEVKAT G+T LGGEDFDN +
Sbjct: 666 -EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNAL 803
Score = 26.9 bits (58), Expect(2) = 8e-56
Identities = 11/14 (78%), Positives = 12/14 (85%)
Frame = +1
Query: 10 VGIDLGTTYSCVGV 23
+GIDLGTT SCV V
Sbjct: 49 IGIDLGTTNSCVSV 90
>TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (22%)
Length = 769
Score = 203 bits (516), Expect = 7e-53
Identities = 105/159 (66%), Positives = 122/159 (76%), Gaps = 1/159 (0%)
Frame = +2
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 80 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 253
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ D+ WPFKVI G DKPMI V YKG EK
Sbjct: 254 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKVIPGPADKPMIVVNYKGDEKQF 433
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVP-AYFND 158
AEEISS++L MK+IAEAYL S +KNA YFND
Sbjct: 434 SAEEISSMVLIKMKEIAEAYLGSTIKNASCHPSLLYFND 550
>BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chaperone hsp70 -
rice (fragment), partial (18%)
Length = 398
Score = 172 bits (436), Expect = 1e-43
Identities = 84/125 (67%), Positives = 100/125 (79%)
Frame = -3
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+V FTD +RL+G AA
Sbjct: 369 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 196
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ DI WPFKV+SG +KPMI V YKG++K
Sbjct: 195 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVLSGPAEKPMIQVSYKGEDKQF 16
Query: 121 CAEEI 125
AEEI
Sbjct: 15 AAEEI 1
>TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (36%)
Length = 827
Score = 99.8 bits (247), Expect(2) = 1e-38
Identities = 55/121 (45%), Positives = 79/121 (64%)
Frame = +1
Query: 89 QKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSIILANMKKIAEAYLKSPVKNA 148
++++ PFK++ N V+ GQ+ + +I + +L MK+ AEAYL + A
Sbjct: 490 KREMKMVPFKIVKAPNGNAW--VETNGQQYS--PSQIGAFVLTKMKETAEAYLGKSISKA 657
Query: 149 VVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIAYGLDNKGRCDGERNIFVFDL 208
V+TVPAYFND+QR+AT +AG IAGL+V RIIN+PTAAA++YG++ K E I FDL
Sbjct: 658 VITVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNKK-----EGLIAXFDL 822
Query: 209 G 209
G
Sbjct: 823 G 825
Score = 77.8 bits (190), Expect(2) = 1e-38
Identities = 46/90 (51%), Positives = 55/90 (61%), Gaps = 1/90 (1%)
Frame = +3
Query: 2 ARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDD-QRLVGGAA 60
+R +GIDLGTT SCV V +E KN ++I N EG + TPS VAF + LVG
Sbjct: 231 SRPAGNDVIGIDLGTTNSCVSV-MEGKNP-KVIENSEGARTTPSVVAFNQKGELLVGTPX 404
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQK 90
K QA NP NT+F KRLIGR+F D QK
Sbjct: 405 KRQAVTNPTNTLFGTKRLIGRRFDDAQTQK 494
>BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cytosolic
[imported] - spinach, partial (21%)
Length = 421
Score = 134 bits (338), Expect = 3e-32
Identities = 65/83 (78%), Positives = 73/83 (87%)
Frame = +1
Query: 163 ATMNAGAIAGLNVMRIINDPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKG 222
AT +AG IAGLNVMRIIN+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+
Sbjct: 1 ATKDAGVIAGLNVMRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE 180
Query: 223 DVFEVKATAGNTHLGGEDFDNRM 245
+FEVKATAG+THLGGEDFDNRM
Sbjct: 181 GIFEVKATAGDTHLGGEDFDNRM 249
>TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, partial
(75%)
Length = 1640
Score = 131 bits (330), Expect = 3e-31
Identities = 62/80 (77%), Positives = 71/80 (88%)
Frame = +2
Query: 166 NAGAIAGLNVMRIINDPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVF 225
+AG I+GLNVMRIIN+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +F
Sbjct: 5 DAGVISGLNVMRIINEPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIF 184
Query: 226 EVKATAGNTHLGGEDFDNRM 245
EVKATAG+THLGGEDFDNRM
Sbjct: 185 EVKATAGDTHLGGEDFDNRM 244
>TC208940 UP|Q39803 (Q39803) BiP isoform C (Fragment), partial (26%)
Length = 447
Score = 90.9 bits (224), Expect(2) = 4e-27
Identities = 41/61 (67%), Positives = 52/61 (85%)
Frame = +1
Query: 30 RVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQNTVFDAKRLIGRKFSDPVVQ 89
+VEII ND+GN+ITPS+VAFTD +RL+G AAK+QAA+NP+ T+FD KRLIGRKF D +
Sbjct: 82 QVEIIANDQGNRITPSWVAFTDSERLIGEAAKNQAAVNPERTIFDVKRLIGRKFEDKEIS 261
Query: 90 K 90
K
Sbjct: 262 K 264
Score = 47.8 bits (112), Expect(2) = 4e-27
Identities = 27/63 (42%), Positives = 41/63 (64%), Gaps = 1/63 (1%)
Frame = +2
Query: 89 QKDIMFWPFKVISGLNDKPMIAVKYK-GQEKNLCAEEISSIILANMKKIAEAYLKSPVKN 147
QKD+ P+K+++ + KP I VK K G+ K EEIS+++L MK+ AEA+L + +
Sbjct: 260 QKDMKLVPYKIVNK-DGKPYIQVKIKDGETKVFSPEEISAMVLIKMKETAEAFLGKKIND 436
Query: 148 AVV 150
AVV
Sbjct: 437 AVV 445
>TC224152 similar to UP|Q8L8N8 (Q8L8N8) 70kD heat shock protein, partial
(33%)
Length = 563
Score = 97.8 bits (242), Expect = 4e-21
Identities = 64/182 (35%), Positives = 101/182 (55%), Gaps = 6/182 (3%)
Frame = +1
Query: 9 AVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINP 68
A+GID+GT+ V VW ++VE++ N KI S+V F D+ + G Q +
Sbjct: 34 AIGIDIGTSQCSVAVW--NGSQVELLKNTRNQKIMKSYVTFKDN--IPSGGVSSQLSHED 201
Query: 69 QN----TVFDAKRLIGRKFSDPVVQ--KDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCA 122
+ T+F+ KRLIGR +DPVV K++ F + G+ +P IA ++
Sbjct: 202 EMLSGATIFNMKRLIGRVDTDPVVHACKNLPFLVQTLDIGV--RPFIAALVNNMWRSTTP 375
Query: 123 EEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDP 182
EE+ +I L ++ +AEA LK ++N V+TVP F+ Q A A+AGL+V+R++ +P
Sbjct: 376 EEVLAIFLVELRAMAEAQLKRRIRNVVLTVPVSFSRFQLTRIERACAMAGLHVLRLMPEP 555
Query: 183 TA 184
TA
Sbjct: 556 TA 561
>BM108096
Length = 525
Score = 94.7 bits (234), Expect = 4e-20
Identities = 46/129 (35%), Positives = 76/129 (58%), Gaps = 1/129 (0%)
Frame = +3
Query: 31 VEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQNTVFDAKRLIGRKFSDPVVQK 90
++++ NDE + TP+ V F D QR +G A +NP+N++ KRLIGR+FSDP +Q+
Sbjct: 33 IDVVLNDESKRETPAIVCFGDKQRFLGTAGAASTMMNPKNSISQIKRLIGRQFSDPELQR 212
Query: 91 DIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSIILANMKKIAEAYLK-SPVKNAV 149
D+ +PF V G + P+I +Y G+ + ++ ++L+N+K+IA L+ A
Sbjct: 213 DLKTFPFVVTEGPDGYPLIXARYLGEARTFTPTQVFGMMLSNLKEIAXKNLECXRCLIAC 392
Query: 150 VTVPAYFND 158
+P YF D
Sbjct: 393 XGIPLYFTD 419
>TC205201 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (18%)
Length = 467
Score = 73.9 bits (180), Expect = 7e-14
Identities = 43/75 (57%), Positives = 51/75 (67%), Gaps = 1/75 (1%)
Frame = +1
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDD-QRLVGGAAKDQAAINP 68
+GIDLGTT SCV V +E KN ++I N EG + TPS VAF + LVG AK QA NP
Sbjct: 247 IGIDLGTTNSCVSV-MEGKNP-KVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNP 420
Query: 69 QNTVFDAKRLIGRKF 83
NT+F KRLIGR+F
Sbjct: 421 TNTLFGTKRLIGRRF 465
>BU081329
Length = 454
Score = 70.1 bits (170), Expect = 1e-12
Identities = 43/134 (32%), Positives = 75/134 (55%), Gaps = 2/134 (1%)
Frame = +3
Query: 111 VKYKGQEKNLCAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAI 170
+KY + +I +++ A++K IAE + V + V+ VP+YF + QR+A ++A AI
Sbjct: 3 LKYLKEIHAFTPVQIVAMLFAHLKTIAEKDFGTAVSDCVIGVPSYFTNLQRQAYLDAAAI 182
Query: 171 AGLNVMRIINDPTAAAIAYGLDNKGRCDGERNIFV--FDLGGGTFDVSLLTIKGDVFEVK 228
GLN + +I+D TA ++YG+ K +I+V D+G VS+ + ++
Sbjct: 183 VGLNSLLLIHDCTATGLSYGV-YKTDIPNAAHIYVAFVDIGHCDTQVSIAAFQAGQMKIL 359
Query: 229 ATAGNTHLGGEDFD 242
+ A ++ LGG DFD
Sbjct: 360 SHAFDSSLGGRDFD 401
>TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (68%)
Length = 1554
Score = 69.7 bits (169), Expect = 1e-12
Identities = 32/37 (86%), Positives = 35/37 (94%)
Frame = +2
Query: 209 GGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
GGGTFDVSLLTI+ +FEVKATAG+THLGGEDFDNRM
Sbjct: 8 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRM 118
>TC233450
Length = 677
Score = 65.5 bits (158), Expect = 2e-11
Identities = 37/111 (33%), Positives = 62/111 (55%)
Frame = +3
Query: 6 DGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAA 65
D VG D+G +CV + ++ ++++ N E + TP+ V F++ QR+ G A A
Sbjct: 219 DMSVVGFDIGNE-NCVIAVVRQRG-IDVLLNYESKRETPAVVCFSEKQRI*GSAGAASAM 392
Query: 66 INPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQ 116
++ ++T+ KRLIGRKF+DP V+K++ P K G I +KY +
Sbjct: 393 MHIKSTISQIKRLIGRKFADPDVKKELKMLPGKTSEGQEGGISIHLKYSAE 545
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,878,387
Number of Sequences: 63676
Number of extensions: 110198
Number of successful extensions: 452
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 425
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 425
length of query: 259
length of database: 12,639,632
effective HSP length: 95
effective length of query: 164
effective length of database: 6,590,412
effective search space: 1080827568
effective search space used: 1080827568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144722.9


