

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.19 + phase: 0
(628 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
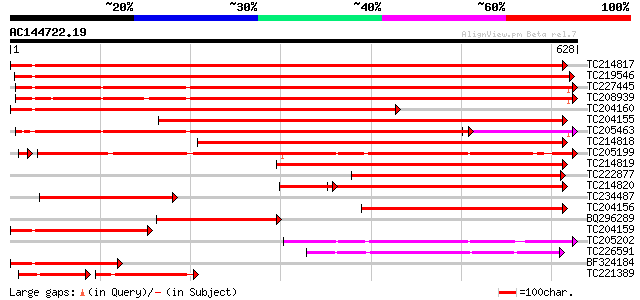
Score E
Sequences producing significant alignments: (bits) Value
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 798 0.0
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 766 0.0
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 618 e-177
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 597 e-171
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 589 e-168
TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, ... 551 e-157
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 520 e-147
TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous hea... 495 e-140
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 426 e-121
TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat sh... 366 e-101
TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa p... 277 1e-74
TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight h... 271 5e-73
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 243 2e-64
TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 223 2e-58
BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cyto... 213 2e-55
TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 208 7e-54
TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 194 8e-50
TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock ... 178 6e-45
BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chapero... 174 8e-44
TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa p... 106 7e-42
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 798 bits (2062), Expect = 0.0
Identities = 407/621 (65%), Positives = 499/621 (79%), Gaps = 3/621 (0%)
Frame = +2
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS V FT+++RLIGDAA
Sbjct: 86 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 259
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP NT+FD KRLIGR++SDS VQ D LWPFKVI GA +KP I+V Y G+EK+F
Sbjct: 260 KNQVAMNPINTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIPGAADKPMIVVNYKGEEKQF 439
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KMREIAEA+L S VKNAV+TVPAYFNDSQR+ATKDAG IAGLNVMRIIN
Sbjct: 440 AAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 619
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 620 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 799
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+ HFV++FK+KN +DIS NP+ALRRLRTACERAKRTLS + TIE+D++Y+GID
Sbjct: 800 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGID 979
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F S++TRA+FEE+NM+LF KCME V CL DAKM+K SVDDVVLVGGS+RIPKV+QLLQ+
Sbjct: 980 FYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQLLQD 1159
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G +M+V
Sbjct: 1160FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1339
Query: 420 VIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHP 478
+IPRNT+IP K+ + T D+Q VLI+V+EGER + DNNLLG F LS +P APRG P
Sbjct: 1340LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTRDNNLLGKFELSGIPPAPRGVP 1519
Query: 479 H-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKF 537
VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ IE+M+QEAE +K++D +
Sbjct: 1520QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEH 1699
Query: 538 KKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
KKKVEA+NAL++Y Y +R +K+D KL P DK+KI A+ + + +Q + F
Sbjct: 1700KKKVEAKNALENYAYNMRNTVKDDKIGEKLDPADKKKIEDAIEQAIQWLDSNQLAEADEF 1879
Query: 598 VDFLKELESIFESAMNKINKG 618
D +KELESI + K+ +G
Sbjct: 1880EDKMKELESICNPIIAKMYQG 1942
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 766 bits (1979), Expect = 0.0
Identities = 391/623 (62%), Positives = 491/623 (78%), Gaps = 3/623 (0%)
Frame = +1
Query: 6 EGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAA 65
EG AIGIDLGTTYSCVGVWQ NDRVEII NDQGNRTTPS VAFT+++RLIGDAAKNQ A
Sbjct: 13 EGKAIGIDLGTTYSCVGVWQ--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVA 186
Query: 66 TNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEI 125
NP NT+FD KRLIGR++SDS VQ D LWPFKV +KP I+V Y G+EK+F AEEI
Sbjct: 187 MNPQNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCDKPMIVVNYKGEEKKFSAEEI 366
Query: 126 SSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
SS++L KMRE+AEAFL VKNAV+TVPAYFNDSQR+ATKDAG I+GLNV+RIINEPTAA
Sbjct: 367 SSMVLVKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAA 546
Query: 186 ALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRM 245
A+AYGL K+A+ ++N+ IFDLGGGTFDVSILT+++ +EVKATAGDTHLGGEDFDNRM
Sbjct: 547 AIAYGLDKKASRKGEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGEDFDNRM 726
Query: 246 LTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSI 305
+ HFV +FK+KN +DIS N +ALRRLRTACERAKRTLS + TIE+D++Y+GIDF ++I
Sbjct: 727 VNHFVSEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYATI 906
Query: 306 TRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGK 365
TRA+FEEMNM+LF KCME V CL DAK++K+ V +VVLVGGS+RIPKV QLLQ+FF GK
Sbjct: 907 TRARFEEMNMDLFRKCMEPVEKCLRDAKIDKSQVHEVVLVGGSTRIPKVHQLLQDFFNGK 1086
Query: 366 ELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
ELC SINPDEAVAYGAA+Q A+L + + V +L+L DVTPLSLG+ G +M+V+IPRN
Sbjct: 1087ELCKSINPDEAVAYGAAVQAAILSGQGDEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRN 1266
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVC 482
T+IP K+ + T D+Q VLI+V+EGER + DNNLLG F L+ +P APRG P VC
Sbjct: 1267TTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVPQINVC 1446
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE+M+++AE +KA+D + KKKVE
Sbjct: 1447FDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAERYKAEDEEVKKKVE 1626
Query: 543 ARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFLK 602
A+N+L++Y Y +R +K++ KL+P +K+KI A+ + +Q + F D K
Sbjct: 1627AKNSLENYAYNMRNTIKDEKIGGKLSPDEKQKIEKAVEDAIQWLEGNQMAEVDEFEDKQK 1806
Query: 603 ELESIFESAMNKINKGDSDEKSD 625
ELE I + K+ +G + D
Sbjct: 1807ELEGICNPIIAKMYQGAAGPGGD 1875
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 618 bits (1593), Expect = e-177
Identities = 329/636 (51%), Positives = 454/636 (70%), Gaps = 14/636 (2%)
Frame = +2
Query: 7 GVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV++ N VEII NDQGNR TPS VAFT+S+RLIG+AAKN AA
Sbjct: 209 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVAFTDSERLIGEAAKNLAAV 382
Query: 67 NPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYM-GKEKRFVAEEI 125
NP TIFDVKRLIGRK+ D VQ D L P+K+++ + KP I VK G+ K F EEI
Sbjct: 383 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVN-KDGKPYIQVKIKDGETKVFSPEEI 559
Query: 126 SSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
S++IL+KM+E AEAFL + +AV+TVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 560 SAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 739
Query: 186 ALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRM 245
A+AYGL K+ ++NI +FDLGGGTFDVSILT+ + +EV AT GDTHLGGEDFD R+
Sbjct: 740 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 910
Query: 246 LTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSI 305
+ +F+K KKK+ +DIS++ +AL +LR ERAKR LS + +E+++++ G+DFS +
Sbjct: 911 MEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEPL 1090
Query: 306 TRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGK 365
TRA+FEE+N +LF K M V + DA ++K+ +D++VLVGGS+RIPKV+QLL+++F GK
Sbjct: 1091TRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1270
Query: 366 ELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
E +NPDEAVAYGAA+Q ++L E + +++L DV PL+LG+ G +M+ +IPRN
Sbjct: 1271EPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1450
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVC 482
T IP K++ + T +D Q+TV I+V+EGER D LLG F LS +P APRG P +V
Sbjct: 1451TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPRGTPQIEVT 1630
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F +DA+GILNV A ++ +G +ITITNEKGRLS+ +I+RM++EAE F +D K K++++
Sbjct: 1631FEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKERID 1810
Query: 543 ARNALDDYLYKVR-KVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFL 601
ARN+L+ Y+Y ++ +V D + KL +KEKI +A+ + + D+Q + + + L
Sbjct: 1811ARNSLETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSVEKEDYEEKL 1990
Query: 602 KELESIFESAMNKINK---------GDSDEKSDSDS 628
KE+E++ ++ + + G S E+ DS
Sbjct: 1991KEVEAVCNPIISAVYQRSGGAPGGGGASGEEDKDDS 2098
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 597 bits (1540), Expect = e-171
Identities = 323/635 (50%), Positives = 446/635 (69%), Gaps = 13/635 (2%)
Frame = +3
Query: 7 GVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV++ N VEII NDQGNR TPS +FT+S+RLIG+AAKN AA
Sbjct: 171 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSW-SFTDSERLIGEAAKNLAAV 341
Query: 67 NPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYM-GKEKRFVAEEI 125
NP IFDVKRLIGRK+ D VQ D L P+K+++ + KP I K G+ K F EEI
Sbjct: 342 NPERVIFDVKRLIGRKFEDKEVQRDMKLVPYKIVN-KDGKPYIQEKIKDGETKVFSPEEI 518
Query: 126 SSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
S++IL+KM+E AEAFL + +AV AYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 519 SAMILTKMKETAEAFLGKKINDAV----AYFNDAQRQATKDAGVIAGLNVARIINEPTAA 686
Query: 186 ALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRM 245
A+AYGL K+ ++NI +FDLGGGTFDVSILT+ + +EV AT GDTHLGGEDFD R+
Sbjct: 687 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 857
Query: 246 LTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSI 305
+ +F+K KK+ +DIS++ +AL +LR ERAKR LS + +E+++++ G+DFS +
Sbjct: 858 MEYFIKLINKKHKKDISKDSRALSKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEPL 1037
Query: 306 TRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGK 365
TRA+FEE+N +LF K M V + DA ++KN +D++VLVGGS+RIPKV+QLL+++F GK
Sbjct: 1038TRARFEELNNDLFRKTMGPVKKAMEDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1217
Query: 366 ELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
E +NPDEAVAYGAA+Q ++L E + +++L DV PL+LG+ G +M+ +IPRN
Sbjct: 1218EPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1397
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVC 482
T IP K++ + T +D Q+TV I+V+EGER D LLG F LS +P APRG P +V
Sbjct: 1398TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFELSGIPPAPRGTPQIEVT 1577
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F +DA+GILNV A ++ +G +ITITNEKGRLS+ +IERM++EAE F +D K K++++
Sbjct: 1578FEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEEDKKVKERID 1757
Query: 543 ARNALDDYLYKVR-KVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFL 601
ARN+L+ Y+Y ++ +V D + KL +KEK+ +A+ + + D+Q + + + L
Sbjct: 1758ARNSLETYVYNMKNQVSDKDKLADKLESDEKEKVETAVKEALEWLDDNQSVEKEEYEEKL 1937
Query: 602 KELESIFESAMNKINK--------GDSDEKSDSDS 628
KE+E++ ++ + + G S E D DS
Sbjct: 1938KEVEAVCNPIISAVYQRSGGAPGGGASGEDDDEDS 2042
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 589 bits (1519), Expect = e-168
Identities = 295/434 (67%), Positives = 360/434 (81%), Gaps = 1/434 (0%)
Frame = +2
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+++RLIGDAA
Sbjct: 95 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 268
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP+NT+FD KRLIGR++SD+ VQ D LWPFKVI G KP I+V Y G+EK+F
Sbjct: 269 KNQVAMNPTNTVFDAKRLIGRRFSDASVQGDMKLWPFKVIPGPAEKPMIVVNYKGEEKQF 448
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KM+EIAEA+L S +KNAV+TVPAYFNDSQR+ATKDAG I+GLNVMRIIN
Sbjct: 449 SAEEISSMVLMKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 628
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 629 EPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 808
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+ HFV++FK+KN +DIS N +ALRRLRTACERAKRTLS + TIE+D++Y+GID
Sbjct: 809 FDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGID 988
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F ++ITRA+FEE+NM+LF KCME V CL DAKM+K++V DVVLVGGS+RIPKV+QLLQ+
Sbjct: 989 FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 1168
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVT LS + +M+V
Sbjct: 1169FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTTLSTALETARGVMTV 1348
Query: 420 VIPRNTSIPVKRTN 433
++ NT+I + +
Sbjct: 1349LMTSNTTISTENAH 1390
>TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, partial
(75%)
Length = 1640
Score = 551 bits (1419), Expect = e-157
Identities = 280/457 (61%), Positives = 361/457 (78%), Gaps = 3/457 (0%)
Frame = +2
Query: 165 KDAGEIAGLNVMRIINEPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNN 224
KDAG I+GLNVMRIINEPTAAA+AYGL K+A ++N+ IFDLGGGTFDVS+LT+++
Sbjct: 2 KDAGVISGLNVMRIINEPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGI 181
Query: 225 YEVKATAGDTHLGGEDFDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSY 284
+EVKATAGDTHLGGEDFDNRM+ HFV++FK+KN +DIS N +ALRRLRTACERAKRTLS
Sbjct: 182 FEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSS 361
Query: 285 DTEATIELDAIYKGIDFSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVL 344
+ TIE+D++Y+GIDF ++ITRA+FEE+NM+LF KCME V CL DAKM+K++V DVVL
Sbjct: 362 TAQTTIEIDSLYEGIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVL 541
Query: 345 VGGSSRIPKVRQLLQEFFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDV 403
VGGS+RIPKV+QLLQ+FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DV
Sbjct: 542 VGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDV 721
Query: 404 TPLSLGVSIRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLL 463
TPLS G+ G +M+V+IPRNT+IP K+ + T D+Q VLI+VYEGER + DNNLL
Sbjct: 722 TPLSTGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLL 901
Query: 464 GLFSLS-LPLAPRGHPH-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIE 521
G F LS +P APRG P VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE
Sbjct: 902 GKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIE 1081
Query: 522 RMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTK 581
+M+QEAE +K++D + KKKVEA+NAL++Y Y +R +K+D +SKL+ DK+KI A+ +
Sbjct: 1082KMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDDKIASKLSSDDKKKIEDAIEQ 1261
Query: 582 GKSLIGDSQQEDTFVFVDFLKELESIFESAMNKINKG 618
+ +Q + F D +KELESI + K+ +G
Sbjct: 1262AIQWLDGNQLAEADEFEDKMKELESICNPIIAKMYQG 1372
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 520 bits (1338), Expect = e-147
Identities = 284/513 (55%), Positives = 372/513 (72%), Gaps = 6/513 (1%)
Frame = +3
Query: 7 GVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDL TTYSCVGV D VEII NDQGNR TPS VAFT+S+RLIG+AAK AA
Sbjct: 192 GTVIGIDL-TTYSCVGV----TDAVEIIANDQGNRITPSWVAFTDSERLIGEAAKIVAAV 356
Query: 67 NPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYM-GKEKRFVAEEI 125
NP TIFDVKRLIGRK+ D VQ D L P+K+++ + KP I VK G+ K F EEI
Sbjct: 357 NPVRTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVN-KDGKPYIQVKIKDGETKVFSPEEI 533
Query: 126 SSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
S++IL+KM+E AEAFL + +AV+TVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 534 SAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 713
Query: 186 ALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRM 245
A+AYGL K+ ++NI +FDLGGGTFDVSILT+ + +EV AT GDTHLGGEDFD R+
Sbjct: 714 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 884
Query: 246 LTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSI 305
+ +F+K KKK+ +DIS++ +AL +LR ERAKR LS + +E+++++ G+DFS +
Sbjct: 885 MEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEPL 1064
Query: 306 TRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGK 365
TRA+FEE+N +LF K M V + DA ++K+ +D++VLVGGS+RIPKV+QLL+++F GK
Sbjct: 1065TRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1244
Query: 366 ELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
E +NPDEAVAYGAA+Q ++L E + +++L DV PL+LG+ G +M+ +IPRN
Sbjct: 1245EPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1424
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVC 482
T IP K++ + T +D Q+TV I+V+EGER D LLG F LS +P APRG P +V
Sbjct: 1425TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPRGTPQIEVT 1604
Query: 483 FTIDAD-GILNVSAVE-ETSGNKNEITITNEKG 513
F +DA+ GILNV A + +G +IT KG
Sbjct: 1605FEVDAERGILNVKARRAKGTGKSEKITDYKRKG 1703
Score = 85.5 bits (210), Expect = 6e-17
Identities = 46/137 (33%), Positives = 82/137 (59%), Gaps = 10/137 (7%)
Frame = +1
Query: 502 NKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVR-KVMKN 560
N+ ITNEKGRLS+ +IERM++EAE F +D K K++++ARN+L+ Y+Y ++ ++
Sbjct: 1669 NQKRSPITNEKGRLSQEEIERMVREAEEFAEEDKKVKERIDARNSLETYVYNMKNQISDK 1848
Query: 561 DSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFLKELESIFESAMNKINK--- 617
D + KL +KEKI +A+ + + D+Q + + + LKE+E++ ++ + +
Sbjct: 1849 DKLADKLESDEKEKIETAVKEALEWLDDNQSMEKEDYEEKLKEVEAVCNPIISAVYQRSG 2028
Query: 618 ------GDSDEKSDSDS 628
G S E+ + DS
Sbjct: 2029 GAPGGGGASGEEDEDDS 2079
>TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (68%)
Length = 1554
Score = 495 bits (1275), Expect = e-140
Identities = 251/413 (60%), Positives = 323/413 (77%), Gaps = 3/413 (0%)
Frame = +2
Query: 209 GGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTHFVKKFKKKNNEDISRNPKAL 268
GGGTFDVS+LT+++ +EVKATAGDTHLGGEDFDNRM+ HFV++FK+KN +DIS NP+AL
Sbjct: 8 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRAL 187
Query: 269 RRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSITRAKFEEMNMNLFEKCMETVNSC 328
RRLRTACERAKRTLS + TIE+D++Y+GIDF S+ITRA+FEE+NM+LF KCME V C
Sbjct: 188 RRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYSTITRARFEELNMDLFRKCMEPVEKC 367
Query: 329 LADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKELCNSINPDEAVAYGAAIQ-AVL 387
L DAKM+K +V DVVLVGGS+RIPKV+QLLQ+FF GKELC SINPDEAVAYGAA+Q A+L
Sbjct: 368 LRDAKMDKRTVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAIL 547
Query: 388 HSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVLI 447
E + V +L+L DVTPLSLG+ G +M+V+IPRNT+IP K+ + T D+Q VLI
Sbjct: 548 SGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLI 727
Query: 448 KVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVCFTIDADGILNVSAVEETSGNKNE 505
+VYEGER + DNNLLG F LS +P APRG P VCF IDA+GILNVSA ++T+G KN+
Sbjct: 728 QVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNK 907
Query: 506 ITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKVMKNDSTSS 565
ITITN+KGRLS+ +IE+M+QEAE +K++D + KKKVEA+NAL++Y Y +R +K++
Sbjct: 908 ITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKKKVEAKNALENYSYNMRNTIKDEKIGG 1087
Query: 566 KLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFLKELESIFESAMNKINKG 618
KL P DK+KI A+ + + +Q + F D +KELESI + K+ +G
Sbjct: 1088KLDPADKKKIEDAIEQAIQWLDSNQLGEADEFEDKMKELESICNPIIAKMYQG 1246
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 426 bits (1095), Expect(2) = e-121
Identities = 255/606 (42%), Positives = 367/606 (60%), Gaps = 9/606 (1%)
Frame = +3
Query: 32 EIIHNDQGNRTTPSCVAFTN-SQRLIGDAAKNQAATNPSNTIFDVKRLIGRKYSDSIVQM 90
++I N +G RTTPS VAF ++ L+G AK QA TNP+NT+F KRLIGR++ DS Q
Sbjct: 186 KVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTKRLIGRRFDDSQTQK 365
Query: 91 DRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSVILSKMREIAEAFLESPVKNAVI 150
+ + P+K++ N + +++ ++ + +L+KM+E AE++L V AVI
Sbjct: 366 EMKMVPYKIVKAPNGDAWV----EANGQQYSPSQVGAFVLTKMKETAESYLGKSVSKAVI 533
Query: 151 TVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALAYGLQKRANFVDKRNIFIFDLGG 210
TVPAYFND+QR+ATKDAG IAGL+V RIINEPTAAAL+YG+ + I +FDLGG
Sbjct: 534 TVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNNKEGL-----IAVFDLGG 698
Query: 211 GTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTHFVKKFKKKNNEDISRNPKALRR 270
GTFDVSIL + + +EVKAT GDT LGGEDFDN +L V +FK+ N D+S++ AL+R
Sbjct: 699 GTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRTENIDLSKDKLALQR 878
Query: 271 LRTACERAKRTLSYDTEATIELDAIYKGID----FSSSITRAKFEEMNMNLFEKCMETVN 326
LR A E+AK LS ++ I L I + ++TR+KFE + +L E+
Sbjct: 879 LREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNHLIERTKAPCK 1058
Query: 327 SCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKELCNSINPDEAVAYGAAIQAV 386
SCL DA + VD+V+LVGG +R+PKV++++ F GK +NPDEAVA GAAIQ
Sbjct: 1059SCLKDANVSIKEVDEVLLVGGMTRVPKVQEVVSAIF-GKSPSKGVNPDEAVAMGAAIQGG 1235
Query: 387 LHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVL 446
+ VK L+L DVTPLSLG+ G I + +I RNT+IP K++ + T D+Q+ V
Sbjct: 1236ILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVG 1406
Query: 447 IKVYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-KVCFTIDADGILNVSAVEETSGNKN 504
IKV +GER DN LG F L +P APRG P +V F IDA+GI+ VSA ++++G +
Sbjct: 1407IKVLQGEREMAVDNKSLGEFELVGIPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQ 1586
Query: 505 EITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKVM--KNDS 562
+ITI G LS +I++M++EAE +D + K ++ RN+ D +Y + K + D
Sbjct: 1587QITI-RSSGGLSEDEIDKMVKEAELHAQKDQERKALIDIRNSADTSIYSIEKSLGEYRDK 1763
Query: 563 TSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFLKELESIFESAMNKINKGDSDE 622
S++ ++ ++ T ++ GD+ E +K A++KI + S
Sbjct: 1764IPSEVAKEIEDAVSDLRT---AMAGDNADE--------IKAKLDAANKAVSKIGEHMSGG 1910
Query: 623 KSDSDS 628
S S S
Sbjct: 1911SSGSSS 1928
Score = 27.7 bits (60), Expect(2) = e-121
Identities = 12/16 (75%), Positives = 13/16 (81%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQ 25
IGIDLGTT SCV V +
Sbjct: 49 IGIDLGTTNSCVSVME 96
>TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat shock cognate
protein 70, partial (54%)
Length = 1266
Score = 366 bits (939), Expect = e-101
Identities = 189/326 (57%), Positives = 245/326 (74%), Gaps = 3/326 (0%)
Frame = +1
Query: 296 YKGIDFSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVR 355
Y+GIDF S++TRA+FEE+NM+LF KCME V CL DAKM+K SVDDVVLVGGS+RIPKV+
Sbjct: 1 YEGIDFYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQ 180
Query: 356 QLLQEFFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRG 414
QLLQ+FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G
Sbjct: 181 QLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAG 360
Query: 415 DIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLA 473
+M+V+IPRNT+IP K+ + T D+Q VLI+V+EGER + DNNLLG F LS +P A
Sbjct: 361 GVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELSGIPPA 540
Query: 474 PRGHPH-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKA 532
PRG P VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ IE+M+QEAE +K+
Sbjct: 541 PRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKS 720
Query: 533 QDMKFKKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQE 592
+D + KKKVEA+NAL++Y Y +R +K+D KL P DK+KI A+ + + +Q
Sbjct: 721 EDEEHKKKVEAKNALENYAYNMRNTVKDDKIGEKLDPTDKKKIEDAIEQAIQWLDSNQLA 900
Query: 593 DTFVFVDFLKELESIFESAMNKINKG 618
+ F D +KELESI + K+ +G
Sbjct: 901 EADEFEDKMKELESICNPIIAKMYQG 978
>TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa protein
(Fragment), partial (19%)
Length = 809
Score = 277 bits (708), Expect = 1e-74
Identities = 148/241 (61%), Positives = 186/241 (76%), Gaps = 4/241 (1%)
Frame = +1
Query: 379 YGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRNTSIPVKRTNEYVTT 438
YGAA+QA L S+ + +VPNLVL D+TPLSLGVS++GD+MSVVIPRNT+IPV+RT YVTT
Sbjct: 1 YGAAVQAALLSKGIVNVPNLVLLDITPLSLGVSVQGDLMSVVIPRNTTIPVRRTKTYVTT 180
Query: 439 EDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPHKVCFTIDADGILNVSAVE 497
ED+QS V+I+VYEGER + SDNNLLG F+LS +P APRGHP F ID +GIL+VSA E
Sbjct: 181 EDNQSAVMIEVYEGERTRASDNNLLGFFTLSGIPPAPRGHPLYETFDIDENGILSVSAEE 360
Query: 498 ETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKV 557
E++GNKNEITITNEK RLS +I+RMIQEAE +KA+D KF +K +A N LD Y+YK++
Sbjct: 361 ESTGNKNEITITNEKERLSTKEIKRMIQEAEYYKAEDKKFLRKAKAMNDLDYYVYKIKNA 540
Query: 558 MKNDSTSSKLTPVDKEKINSAMTKGKSLIGD-SQQEDTFVFVDFLKELESIFE--SAMNK 614
+K SSKL +KE ++SA+ + L+ D +QQ+D VF D LKELESI E AM K
Sbjct: 541 LKKKDISSKLCSKEKENVSSAIARATDLLEDNNQQDDIVVFEDNLKELESIIERMKAMGK 720
Query: 615 I 615
I
Sbjct: 721 I 723
>TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight heat shock
protein, partial (54%)
Length = 1272
Score = 271 bits (694), Expect = 5e-73
Identities = 144/269 (53%), Positives = 196/269 (72%), Gaps = 3/269 (1%)
Frame = +2
Query: 353 KVRQLLQEFFQGKELCNSINPDEAVAYGAAIQA-VLHSESVKSVPNLVLRDVTPLSLGVS 411
K +QL+Q+F+ GKELC SINPDEAVAYGAA+QA +L E + V +L+L DVTPLSLG+
Sbjct: 164 K*QQLVQDFYNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLE 343
Query: 412 IRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-L 470
G +M+V+IPRNT+IP K+ + T D+Q VLI+VYEGER + DNNLLG F LS +
Sbjct: 344 TAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGI 523
Query: 471 PLAPRGHPH-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAEN 529
P APRG P VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE+M+QEAE
Sbjct: 524 PPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQEAEK 703
Query: 530 FKAQDMKFKKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDS 589
+KA+D + KKKV+A+NAL++Y Y +R +K++ +SKL+ DK+KI A+ + +
Sbjct: 704 YKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIASKLSDDDKKKIEDAIESAIQWLDGN 883
Query: 590 QQEDTFVFVDFLKELESIFESAMNKINKG 618
Q + F D +KELESI + K+ +G
Sbjct: 884 QLAEADEFEDKMKELESICNPIIAKMYQG 970
Score = 87.0 bits (214), Expect = 2e-17
Identities = 41/65 (63%), Positives = 52/65 (79%)
Frame = +1
Query: 299 IDFSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLL 358
+DF ++ITRA+FEE+NM+LF KCME V CL DAKM+K++V DVVLVGGS+RIPKV +
Sbjct: 1 VDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVAAIG 180
Query: 359 QEFFQ 363
F Q
Sbjct: 181 AGFLQ 195
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 243 bits (619), Expect = 2e-64
Identities = 114/152 (75%), Positives = 136/152 (89%)
Frame = +1
Query: 34 IHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRL 93
IHNDQGN TTPSCVAFT+ QRLIG+AAKNQAATNP NT+FD KRLIGRK+SD ++Q D++
Sbjct: 1 IHNDQGNNTTPSCVAFTDQQRLIGEAAKNQAATNPENTVFDAKRLIGRKFSDPVIQKDKM 180
Query: 94 LWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSVILSKMREIAEAFLESPVKNAVITVP 153
LWPFKV++G N+KP I + Y G+EK +AEE+SS++L KMREIAEA+LE+PV+NAV+TVP
Sbjct: 181 LWPFKVVAGINDKPMISLNYKGQEKHLLAEEVSSMVLIKMREIAEAYLETPVENAVVTVP 360
Query: 154 AYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
AYFNDSQR+AT DAG IAGLNVMRIINEPTAA
Sbjct: 361 AYFNDSQRKATIDAGAIAGLNVMRIINEPTAA 456
>TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (40%)
Length = 965
Score = 223 bits (569), Expect = 2e-58
Identities = 119/231 (51%), Positives = 164/231 (70%), Gaps = 2/231 (0%)
Frame = +2
Query: 390 ESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVLIKV 449
E + V +L+L DVTPLSLG+ G +M+V+IPRNT+IP K+ + T D+Q VLI+V
Sbjct: 2 EGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQV 181
Query: 450 YEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVCFTIDADGILNVSAVEETSGNKNEIT 507
YEGER + DNNLLG F LS +P APRG P VCF IDA+GILNVSA ++T+G KN+IT
Sbjct: 182 YEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKIT 361
Query: 508 ITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKVMKNDSTSSKL 567
ITN+KGRLS+ +IE+M+QEAE +KA+D + KKKVEA+N L++Y Y +R +K+D +SKL
Sbjct: 362 ITNDKGRLSKEEIEKMVQEAEKYKAEDEEHKKKVEAKNTLENYAYNMRNTIKDDKIASKL 541
Query: 568 TPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFLKELESIFESAMNKINKG 618
+ DK+KI A+ + + +Q + F D +KELESI + K+ +G
Sbjct: 542 SADDKKKIEDAIEQAIQWLDGNQLAEADEFEDKMKELESICNPIIAKMYQG 694
>BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cytosolic
[imported] - spinach, partial (21%)
Length = 421
Score = 213 bits (543), Expect = 2e-55
Identities = 100/139 (71%), Positives = 124/139 (88%)
Frame = +1
Query: 163 ATKDAGEIAGLNVMRIINEPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKD 222
ATKDAG IAGLNVMRIINEPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT+++
Sbjct: 1 ATKDAGVIAGLNVMRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE 180
Query: 223 NNYEVKATAGDTHLGGEDFDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTL 282
+EVKATAGDTHLGGEDFDNRM+ HFV++FK+K+ +DI+ NP+ALRRLRTACERAKRTL
Sbjct: 181 GIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKHKKDINGNPRALRRLRTACERAKRTL 360
Query: 283 SYDTEATIELDAIYKGIDF 301
S + TIE+D++Y+G+DF
Sbjct: 361 SSTAQTTIEIDSLYEGVDF 417
>TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (22%)
Length = 769
Score = 208 bits (529), Expect = 7e-54
Identities = 109/159 (68%), Positives = 125/159 (78%), Gaps = 1/159 (0%)
Frame = +2
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+++RLIGDAA
Sbjct: 80 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 253
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP NT+FD KRLIGR++SD+ VQ D LWPFKVI G +KP I+V Y G EK+F
Sbjct: 254 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKVIPGPADKPMIVVNYKGDEKQF 433
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVP-AYFND 158
AEEISS++L KM+EIAEA+L S +KNA YFND
Sbjct: 434 SAEEISSMVLIKMKEIAEAYLGSTIKNASCHPSLLYFND 550
>TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (51%)
Length = 1228
Score = 194 bits (494), Expect = 8e-50
Identities = 126/328 (38%), Positives = 190/328 (57%), Gaps = 3/328 (0%)
Frame = +2
Query: 304 SITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQ 363
++TR+KFE + +L E+ SCL DA + VD+V+LVGG +R+PKV++++ E F
Sbjct: 41 TLTRSKFEALVNHLIERTKVPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF- 217
Query: 364 GKELCNSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPR 423
GK +NPDEAVA GAAIQ + VK L+L DVTPLSLG+ G I + +I R
Sbjct: 218 GKSPSKGVNPDEAVAMGAAIQGGILRGDVKE---LLLLDVTPLSLGIETLGGIFTRLINR 388
Query: 424 NTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-KV 481
NT+IP K++ + T D+Q+ V IKV +GER +DN +LG F L +P APRG P +V
Sbjct: 389 NTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMAADNKMLGEFDLVGIPPAPRGLPQIEV 568
Query: 482 CFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKV 541
F IDA+GI+ VSA ++++G + +ITI G LS +IE+M++EAE +D + K +
Sbjct: 569 TFDIDANGIVTVSAKDKSTGKEQQITI-RSSGGLSEDEIEKMVKEAELHAQKDQERKALI 745
Query: 542 EARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKG-KSLIGDSQQEDTFVFVDF 600
+ RN+ D +Y + K + ++KI S + K + + D ++ + VD
Sbjct: 746 DIRNSADTTIYSIEKSLGE----------YRDKIPSEVAKEIEDAVSDLRKAMSEDNVDE 895
Query: 601 LKELESIFESAMNKINKGDSDEKSDSDS 628
+K A++KI + S S S
Sbjct: 896 IKSKLDAANKAVSKIGEHMSGGSSGGSS 979
>TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock protein
hsp70. {Pisum sativum;} , partial (45%)
Length = 1338
Score = 178 bits (452), Expect = 6e-45
Identities = 111/289 (38%), Positives = 172/289 (59%), Gaps = 3/289 (1%)
Frame = +3
Query: 329 LADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKELCNSINPDEAVAYGAAIQAVLH 388
L DAK+ +D+V+LVGGS+RIP V++L+++ GK+ ++NPDE VA GAA+QA +
Sbjct: 3 LRDAKLSFKDLDEVILVGGSTRIPAVQELVKKL-TGKDPNVTVNPDEVVALGAAVQAGVL 179
Query: 389 SESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVLIK 448
+ V + VL DVTPLSLG+ G +M+ +IPRNT++P ++ + T D Q++V I
Sbjct: 180 AGDVSDI---VLLDVTPLSLGLETLGGVMTKIIPRNTTLPTSKSEVFSTAADGQTSVEIN 350
Query: 449 VYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-KVCFTIDADGILNVSAVEETSGNKNEI 506
V +GER DN LG F L +P APRG P +V F IDA+GIL+V+A+++ +G K +I
Sbjct: 351 VLQGEREFVRDNKSLGSFRLDGIPPAPRGVPQIEVKFDIDANGILSVTAIDKGTGKKQDI 530
Query: 507 TITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKVMKNDSTSSK 566
TIT L ++ERM+ EAE F +D + + ++ +N D +Y+ K +K K
Sbjct: 531 TITG-ASTLPSDEVERMVNEAEKFSKEDKEKRDAIDTKNQADSVVYQTEKQLK--ELGDK 701
Query: 567 LTPVDKEKINSAMTKGKSLI-GDSQQEDTFVFVDFLKELESIFESAMNK 614
+ KEK+ + + + K I G S Q +E+ + +S N+
Sbjct: 702 VPGPVKEKVEAKLGELKDAISGGSTQAIKDAMAALNQEVMQLGQSLYNQ 848
>BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chaperone hsp70 -
rice (fragment), partial (18%)
Length = 398
Score = 174 bits (442), Expect = 8e-44
Identities = 88/125 (70%), Positives = 100/125 (79%)
Frame = -3
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS V FT+++RLIGDAA
Sbjct: 369 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 196
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP NT+FD KRLIGR++SD+ VQ D LWPFKV+SG KP I V Y G++K+F
Sbjct: 195 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVLSGPAEKPMIQVSYKGEDKQF 16
Query: 121 VAEEI 125
AEEI
Sbjct: 15 AAEEI 1
>TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (36%)
Length = 827
Score = 106 bits (264), Expect(2) = 7e-42
Identities = 56/114 (49%), Positives = 75/114 (65%)
Frame = +1
Query: 96 PFKVISGANNKPTIIVKYMGKEKRFVAEEISSVILSKMREIAEAFLESPVKNAVITVPAY 155
PFK++ N + +++ +I + +L+KM+E AEA+L + AVITVPAY
Sbjct: 511 PFKIVKAPNGNAWVETN----GQQYSPSQIGAFVLTKMKETAEAYLGKSISKAVITVPAY 678
Query: 156 FNDSQRRATKDAGEIAGLNVMRIINEPTAAALAYGLQKRANFVDKRNIFIFDLG 209
FND+QR+ATKDAG IAGL+V RIINEPTAAAL+YG+ K+ I FDLG
Sbjct: 679 FNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNKKEGL-----IAXFDLG 825
Score = 83.6 bits (205), Expect(2) = 7e-42
Identities = 44/81 (54%), Positives = 55/81 (67%), Gaps = 1/81 (1%)
Frame = +3
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
IGIDLGTT SCV V + +N +V I N +G RTTPS VAF + L+G K QA TNP
Sbjct: 255 IGIDLGTTNSCVSVMEGKNPKV--IENSEGARTTPSVVAFNQKGELLVGTPXKRQAVTNP 428
Query: 69 SNTIFDVKRLIGRKYSDSIVQ 89
+NT+F KRLIGR++ D+ Q
Sbjct: 429 TNTLFGTKRLIGRRFDDAQTQ 491
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.132 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,746,819
Number of Sequences: 63676
Number of extensions: 235214
Number of successful extensions: 906
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 834
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 839
length of query: 628
length of database: 12,639,632
effective HSP length: 103
effective length of query: 525
effective length of database: 6,081,004
effective search space: 3192527100
effective search space used: 3192527100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144722.19


