

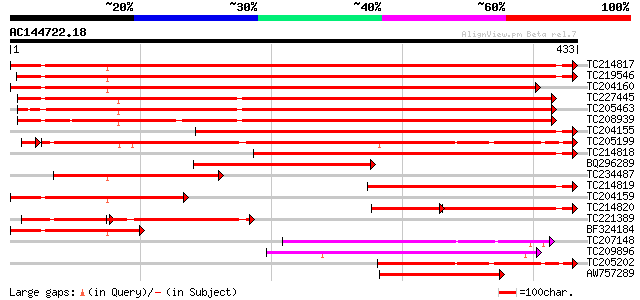
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.18 + phase: 0 /pseudo
(433 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 579 e-166
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 563 e-161
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 549 e-157
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 439 e-123
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 426 e-119
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 421 e-118
TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, ... 379 e-105
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 317 3e-88
TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous hea... 319 2e-87
BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cyto... 210 1e-54
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 201 7e-52
TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat sh... 194 6e-50
TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 170 1e-42
TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight h... 104 1e-41
TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa p... 101 2e-36
BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chapero... 134 6e-32
TC207148 heat shock protein 70-related protein 120 9e-28
TC209896 similar to UP|O23508 (O23508) Growth regulator like pro... 112 3e-25
TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 110 2e-24
AW757289 104 9e-23
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 579 bits (1492), Expect = e-166
Identities = 301/456 (66%), Positives = 360/456 (78%), Gaps = 23/456 (5%)
Frame = +2
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS V FT+++RLIGDAA
Sbjct: 86 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 259
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP NT+F FKVI G DKP IVVNYKGEEK+F
Sbjct: 260 KNQVAMNPINTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIPGAADKPMIVVNYKGEEKQF 439
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KMREIAEA+L + VKNAV++VPAYFNDSQR+ATKDAG IAGLNVM+IIN
Sbjct: 440 AAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 619
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 620 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 799
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV+HFV+EF+ K K+DIS N +ALRRLRTACE+AKRTLS TIE+D++Y+GID
Sbjct: 800 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGID 979
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F S++TRA+FEE+NM+LF KCM V CL DAKMDK SVDDVVLVGGS+RIPKV+QLLQ+
Sbjct: 980 FYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQLLQD 1159
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+ G +M+V
Sbjct: 1160FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1339
Query: 398 MIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+IPRNT+IP E+ + Y N+ V ++V++
Sbjct: 1340LIPRNTTIPTKKEQVFSTYS----DNQPGVLIQVFE 1435
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 563 bits (1452), Expect = e-161
Identities = 294/451 (65%), Positives = 354/451 (78%), Gaps = 23/451 (5%)
Frame = +1
Query: 6 EGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAA 65
EG AIGIDLGTTYSCVGVW QNDRVEII NDQGNRTTPS VAFT+++RLIGDAAKNQ A
Sbjct: 13 EGKAIGIDLGTTYSCVGVW--QNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVA 186
Query: 66 TNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRFVAEEI 103
NP NT+F FKV DKP IVVNYKGEEK+F AEEI
Sbjct: 187 MNPQNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCDKPMIVVNYKGEEKKFSAEEI 366
Query: 104 SSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAA 163
SS++L KMRE+AEAFL VKNAV++VPAYFNDSQR+ATKDAGAI+GLNV++IINEPTAA
Sbjct: 367 SSMVLVKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAA 546
Query: 164 ALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIM 223
A+AYGL K+A+ ++N+ IFDLGGGTFDVSILT++ +FEVKATAGDTHLGG+DFDN M
Sbjct: 547 AIAYGLDKKASRKGEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGEDFDNRM 726
Query: 224 VDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSI 283
V+HFV EF+ K K+DIS N++ALRRLRTACE+AKRTLS TIE+D++Y+GIDF ++I
Sbjct: 727 VNHFVSEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYATI 906
Query: 284 TRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGK 343
TRA+FEEMNM+LF KCM V CL DAK+DK+ V +VVLVGGS+RIPKV QLLQ+ F GK
Sbjct: 907 TRARFEEMNMDLFRKCMEPVEKCLRDAKIDKSQVHEVVLVGGSTRIPKVHQLLQDFFNGK 1086
Query: 344 ELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRN 402
ELCKSINPDEAVAYGA VQAA+LS +G + V +L+L DVTPLS G+ G +M+V+IPRN
Sbjct: 1087ELCKSINPDEAVAYGAAVQAAILSGQGDEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRN 1266
Query: 403 TSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
T+IP E+ + Y N+ V ++V++
Sbjct: 1267TTIPTKKEQIFSTYS----DNQPGVLIQVFE 1347
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 549 bits (1415), Expect = e-157
Identities = 283/428 (66%), Positives = 344/428 (80%), Gaps = 23/428 (5%)
Frame = +2
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+++RLIGDAA
Sbjct: 95 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 268
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP+NT+F FKVI G +KP IVVNYKGEEK+F
Sbjct: 269 KNQVAMNPTNTVFDAKRLIGRRFSDASVQGDMKLWPFKVIPGPAEKPMIVVNYKGEEKQF 448
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KM+EIAEA+L + +KNAV++VPAYFNDSQR+ATKDAG I+GLNVM+IIN
Sbjct: 449 SAEEISSMVLMKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 628
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 629 EPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 808
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV+HFV+EF+ K K+DIS N++ALRRLRTACE+AKRTLS TIE+D++Y+GID
Sbjct: 809 FDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGID 988
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F ++ITRA+FEE+NM+LF KCM V CL DAKMDK++V DVVLVGGS+RIPKV+QLLQ+
Sbjct: 989 FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 1168
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVT LS + +M+V
Sbjct: 1169FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTTLSTALETARGVMTV 1348
Query: 398 MIPRNTSI 405
++ NT+I
Sbjct: 1349LMTSNTTI 1372
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 439 bits (1128), Expect = e-123
Identities = 232/434 (53%), Positives = 307/434 (70%), Gaps = 23/434 (5%)
Frame = +2
Query: 7 GVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV+ +N VEII NDQGNR TPS VAFT+S+RLIG+AAKN AA
Sbjct: 209 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVAFTDSERLIGEAAKNLAAV 382
Query: 67 NPSNTIFAFKVIAGT---------------------NDKPTIVVNYK-GEEKRFVAEEIS 104
NP TIF K + G + KP I V K GE K F EEIS
Sbjct: 383 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKDGKPYIQVKIKDGETKVFSPEEIS 562
Query: 105 SVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAA 164
++IL+KM+E AEAFL + +AV++VPAYFND+QR+ATKDAG IAGLNV +IINEPTAAA
Sbjct: 563 AMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAA 742
Query: 165 LAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMV 224
+AYGL K+ ++NI +FDLGGGTFDVSILT+ N VFEV AT GDTHLGG+DFD ++
Sbjct: 743 IAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIM 913
Query: 225 DHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSIT 284
++F+K + K+ +DIS++++AL +LR E+AKR LS +E+++++ G+DF +T
Sbjct: 914 EYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEPLT 1093
Query: 285 RAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKE 344
RA+FEE+N +LF K M V + DA + K+ +D++VLVGGS+RIPKV+QLL++ F GKE
Sbjct: 1094RARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDGKE 1273
Query: 345 LCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNT 403
K +NPDEAVAYGA VQ ++LS EG + +++L DV PL+ G+ +G +M+ +IPRNT
Sbjct: 1274PNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRNT 1453
Query: 404 SIPVTVEKTKTYYE 417
IP + T Y+
Sbjct: 1454VIPTKKSQVFTTYQ 1495
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 426 bits (1094), Expect = e-119
Identities = 230/434 (52%), Positives = 303/434 (68%), Gaps = 23/434 (5%)
Frame = +3
Query: 7 GVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDL TTYSCVGV D VEII NDQGNR TPS VAFT+S+RLIG+AAK AA
Sbjct: 192 GTVIGIDL-TTYSCVGV----TDAVEIIANDQGNRITPSWVAFTDSERLIGEAAKIVAAV 356
Query: 67 NPSNTIFAFKVIAGT---------------------NDKPTIVVNYK-GEEKRFVAEEIS 104
NP TIF K + G + KP I V K GE K F EEIS
Sbjct: 357 NPVRTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKDGKPYIQVKIKDGETKVFSPEEIS 536
Query: 105 SVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAA 164
++IL+KM+E AEAFL + +AV++VPAYFND+QR+ATKDAG IAGLNV +IINEPTAAA
Sbjct: 537 AMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAA 716
Query: 165 LAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMV 224
+AYGL K+ ++NI +FDLGGGTFDVSILT+ N VFEV AT GDTHLGG+DFD ++
Sbjct: 717 IAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIM 887
Query: 225 DHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSIT 284
++F+K + K+ +DIS++++AL +LR E+AKR LS +E+++++ G+DF +T
Sbjct: 888 EYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEPLT 1067
Query: 285 RAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKE 344
RA+FEE+N +LF K M V + DA + K+ +D++VLVGGS+RIPKV+QLL++ F GKE
Sbjct: 1068RARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDGKE 1247
Query: 345 LCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNT 403
K +NPDEAVAYGA VQ ++LS EG + +++L DV PL+ G+ +G +M+ +IPRNT
Sbjct: 1248PNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRNT 1427
Query: 404 SIPVTVEKTKTYYE 417
IP + T Y+
Sbjct: 1428VIPTKKSQVFTTYQ 1469
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 421 bits (1082), Expect = e-118
Identities = 229/434 (52%), Positives = 300/434 (68%), Gaps = 23/434 (5%)
Frame = +3
Query: 7 GVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV+ +N VEII NDQGNR TPS +FT+S+RLIG+AAKN AA
Sbjct: 171 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSW-SFTDSERLIGEAAKNLAAV 341
Query: 67 NPSNTIFAFKVIAGT---------------------NDKPTIVVNYK-GEEKRFVAEEIS 104
NP IF K + G + KP I K GE K F EEIS
Sbjct: 342 NPERVIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKDGKPYIQEKIKDGETKVFSPEEIS 521
Query: 105 SVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAA 164
++IL+KM+E AEAFL + +AV AYFND+QR+ATKDAG IAGLNV +IINEPTAAA
Sbjct: 522 AMILTKMKETAEAFLGKKINDAV----AYFNDAQRQATKDAGVIAGLNVARIINEPTAAA 689
Query: 165 LAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMV 224
+AYGL K+ ++NI +FDLGGGTFDVSILT+ N VFEV AT GDTHLGG+DFD ++
Sbjct: 690 IAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIM 860
Query: 225 DHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSIT 284
++F+K K+K+DIS++S+AL +LR E+AKR LS +E+++++ G+DF +T
Sbjct: 861 EYFIKLINKKHKKDISKDSRALSKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEPLT 1040
Query: 285 RAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKE 344
RA+FEE+N +LF K M V + DA + KN +D++VLVGGS+RIPKV+QLL++ F GKE
Sbjct: 1041RARFEELNNDLFRKTMGPVKKAMEDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGKE 1220
Query: 345 LCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNT 403
K +NPDEAVAYGA VQ ++LS EG + +++L DV PL+ G+ +G +M+ +IPRNT
Sbjct: 1221PNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRNT 1400
Query: 404 SIPVTVEKTKTYYE 417
IP + T Y+
Sbjct: 1401VIPTKKSQVFTTYQ 1442
>TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, partial
(75%)
Length = 1640
Score = 379 bits (972), Expect = e-105
Identities = 190/292 (65%), Positives = 239/292 (81%), Gaps = 1/292 (0%)
Frame = +2
Query: 143 KDAGAIAGLNVMQIINEPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNV 202
KDAG I+GLNVM+IINEPTAAA+AYGL K+A ++N+ IFDLGGGTFDVS+LT++ +
Sbjct: 2 KDAGVISGLNVMRIINEPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGI 181
Query: 203 FEVKATAGDTHLGGQDFDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSY 262
FEVKATAGDTHLGG+DFDN MV+HFV+EF+ K K+DIS N++ALRRLRTACE+AKRTLS
Sbjct: 182 FEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSS 361
Query: 263 DTDATIELDAIYKGIDFCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVL 322
TIE+D++Y+GIDF ++ITRA+FEE+NM+LF KCM V CL DAKMDK++V DVVL
Sbjct: 362 TAQTTIEIDSLYEGIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVL 541
Query: 323 VGGSSRIPKVRQLLQEVFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDV 381
VGGS+RIPKV+QLLQ+ F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DV
Sbjct: 542 VGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDV 721
Query: 382 TPLSFGVSELGNLMSVMIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
TPLS G+ G +M+V+IPRNT+IP E+ + Y N+ V ++VY+
Sbjct: 722 TPLSTGLETAGGVMTVLIPRNTTIPTKKEQVFSTYS----DNQPGVLIQVYE 865
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 317 bits (811), Expect(2) = 3e-88
Identities = 191/432 (44%), Positives = 262/432 (60%), Gaps = 23/432 (5%)
Frame = +3
Query: 25 HEQNDRVEIIHNDQGNRTTPSCVAFTN-SQRLIGDAAKNQAATNPSNTIFAFKVIAGTN- 82
H QN +V I N +G RTTPS VAF ++ L+G AK QA TNP+NT+F K + G
Sbjct: 171 HFQNPKV--IENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTKRLIGRRF 344
Query: 83 -----DKPTIVVNYK------------GEEKRFVAEEISSVILSKMREIAEAFLQTPVKN 125
K +V YK +++ ++ + +L+KM+E AE++L V
Sbjct: 345 DDSQTQKEMKMVPYKIVKAPNGDAWVEANGQQYSPSQVGAFVLTKMKETAESYLGKSVSK 524
Query: 126 AVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQKRANCVDKRNIFIFD 185
AVI+VPAYFND+QR+ATKDAG IAGL+V +IINEPTAAAL+YG+ + I +FD
Sbjct: 525 AVITVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNNKEGL-----IAVFD 689
Query: 186 LGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKEFRNKYKEDISENSKA 245
LGGGTFDVSIL + N VFEVKAT GDT LGG+DFDN ++D V EF+ D+S++ A
Sbjct: 690 LGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRTENIDLSKDKLA 869
Query: 246 LRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCS----SITRAKFEEMNMNLFEKCMN 301
L+RLR A EKAK LS + I L I ++TR+KFE + +L E+
Sbjct: 870 LQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNHLIERTKA 1049
Query: 302 TVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELCKSINPDEAVAYGAVV 361
CL DA + VD+V+LVGG +R+PKV++++ +F GK K +NPDEAVA GA +
Sbjct: 1050PCKSCLKDANVSIKEVDEVLLVGGMTRVPKVQEVVSAIF-GKSPSKGVNPDEAVAMGAAI 1226
Query: 362 QAALLSEGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSIPVTVEKTKTYYECYDY 421
Q +L +V L+L DVTPLS G+ LG + + +I RNT+IP +K++ + D
Sbjct: 1227QGGIL---RGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIP--TKKSQVFSTAAD- 1388
Query: 422 YNKYSVTVKVYQ 433
N+ V +KV Q
Sbjct: 1389-NQTQVGIKVLQ 1421
Score = 27.3 bits (59), Expect(2) = 3e-88
Identities = 12/14 (85%), Positives = 12/14 (85%)
Frame = +1
Query: 10 IGIDLGTTYSCVGV 23
IGIDLGTT SCV V
Sbjct: 49 IGIDLGTTNSCVSV 90
>TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (68%)
Length = 1554
Score = 319 bits (817), Expect = 2e-87
Identities = 161/248 (64%), Positives = 200/248 (79%), Gaps = 1/248 (0%)
Frame = +2
Query: 187 GGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKEFRNKYKEDISENSKAL 246
GGGTFDVS+LT++ +FEVKATAGDTHLGG+DFDN MV+HFV+EF+ K K+DIS N +AL
Sbjct: 8 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRAL 187
Query: 247 RRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSITRAKFEEMNMNLFEKCMNTVNIC 306
RRLRTACE+AKRTLS TIE+D++Y+GIDF S+ITRA+FEE+NM+LF KCM V C
Sbjct: 188 RRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYSTITRARFEELNMDLFRKCMEPVEKC 367
Query: 307 LADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELCKSINPDEAVAYGAVVQAALL 366
L DAKMDK +V DVVLVGGS+RIPKV+QLLQ+ F GKELCKSINPDEAVAYGA VQAA+L
Sbjct: 368 LRDAKMDKRTVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAIL 547
Query: 367 S-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSIPVTVEKTKTYYECYDYYNKY 425
S EG++ V +L+L DVTPLS G+ G +M+V+IPRNT+IP E+ + Y N+
Sbjct: 548 SGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYS----DNQP 715
Query: 426 SVTVKVYQ 433
V ++VY+
Sbjct: 716 GVLIQVYE 739
>BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cytosolic
[imported] - spinach, partial (21%)
Length = 421
Score = 210 bits (534), Expect = 1e-54
Identities = 98/139 (70%), Positives = 122/139 (87%)
Frame = +1
Query: 141 ATKDAGAIAGLNVMQIINEPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKN 200
ATKDAG IAGLNVM+IINEPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT++
Sbjct: 1 ATKDAGVIAGLNVMRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE 180
Query: 201 NVFEVKATAGDTHLGGQDFDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTL 260
+FEVKATAGDTHLGG+DFDN MV+HFV+EF+ K+K+DI+ N +ALRRLRTACE+AKRTL
Sbjct: 181 GIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKHKKDINGNPRALRRLRTACERAKRTL 360
Query: 261 SYDTDATIELDAIYKGIDF 279
S TIE+D++Y+G+DF
Sbjct: 361 SSTAQTTIEIDSLYEGVDF 417
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 201 bits (510), Expect = 7e-52
Identities = 102/152 (67%), Positives = 119/152 (78%), Gaps = 22/152 (14%)
Frame = +1
Query: 34 IHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPSNTIF-------------------- 73
IHNDQGN TTPSCVAFT+ QRLIG+AAKNQAATNP NT+F
Sbjct: 1 IHNDQGNNTTPSCVAFTDQQRLIGEAAKNQAATNPENTVFDAKRLIGRKFSDPVIQKDKM 180
Query: 74 --AFKVIAGTNDKPTIVVNYKGEEKRFVAEEISSVILSKMREIAEAFLQTPVKNAVISVP 131
FKV+AG NDKP I +NYKG+EK +AEE+SS++L KMREIAEA+L+TPV+NAV++VP
Sbjct: 181 LWPFKVVAGINDKPMISLNYKGQEKHLLAEEVSSMVLIKMREIAEAYLETPVENAVVTVP 360
Query: 132 AYFNDSQRRATKDAGAIAGLNVMQIINEPTAA 163
AYFNDSQR+AT DAGAIAGLNVM+IINEPTAA
Sbjct: 361 AYFNDSQRKATIDAGAIAGLNVMRIINEPTAA 456
>TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat shock cognate
protein 70, partial (54%)
Length = 1266
Score = 194 bits (493), Expect = 6e-50
Identities = 100/161 (62%), Positives = 126/161 (78%), Gaps = 1/161 (0%)
Frame = +1
Query: 274 YKGIDFCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVR 333
Y+GIDF S++TRA+FEE+NM+LF KCM V CL DAKMDK SVDDVVLVGGS+RIPKV+
Sbjct: 1 YEGIDFYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQ 180
Query: 334 QLLQEVFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELG 392
QLLQ+ F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+ G
Sbjct: 181 QLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAG 360
Query: 393 NLMSVMIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+M+V+IPRNT+IP E+ + Y N+ V ++V++
Sbjct: 361 GVMTVLIPRNTTIPTKKEQVFSTYS----DNQPGVLIQVFE 471
>TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (22%)
Length = 769
Score = 170 bits (431), Expect = 1e-42
Identities = 97/159 (61%), Positives = 110/159 (69%), Gaps = 23/159 (14%)
Frame = +2
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+++RLIGDAA
Sbjct: 80 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 253
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP NT+F FKVI G DKP IVVNYKG+EK+F
Sbjct: 254 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKVIPGPADKPMIVVNYKGDEKQF 433
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVP-AYFND 136
AEEISS++L KM+EIAEA+L + +KNA YFND
Sbjct: 434 SAEEISSMVLIKMKEIAEAYLGSTIKNASCHPSLLYFND 550
>TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight heat shock
protein, partial (54%)
Length = 1272
Score = 104 bits (260), Expect(2) = 1e-41
Identities = 56/104 (53%), Positives = 76/104 (72%), Gaps = 1/104 (0%)
Frame = +2
Query: 331 KVRQLLQEVFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVS 389
K +QL+Q+ + GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+
Sbjct: 164 K*QQLVQDFYNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLE 343
Query: 390 ELGNLMSVMIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
G +M+V+IPRNT+IP E+ + Y N+ V ++VY+
Sbjct: 344 TAGGVMTVLIPRNTTIPTKKEQVFSTYS----DNQPGVLIQVYE 463
Score = 84.0 bits (206), Expect(2) = 1e-41
Identities = 39/56 (69%), Positives = 48/56 (85%)
Frame = +1
Query: 277 IDFCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKV 332
+DF ++ITRA+FEE+NM+LF KCM V CL DAKMDK++V DVVLVGGS+RIPKV
Sbjct: 1 VDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKV 168
>TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (36%)
Length = 827
Score = 101 bits (251), Expect(2) = 2e-36
Identities = 54/113 (47%), Positives = 75/113 (65%)
Frame = +1
Query: 75 FKVIAGTNDKPTIVVNYKGEEKRFVAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYF 134
FK++ N + N +++ +I + +L+KM+E AEA+L + AVI+VPAYF
Sbjct: 514 FKIVKAPNGNAWVETN----GQQYSPSQIGAFVLTKMKETAEAYLGKSISKAVITVPAYF 681
Query: 135 NDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQKRANCVDKRNIFIFDLG 187
ND+QR+ATKDAG IAGL+V +IINEPTAAAL+YG+ K+ I FDLG
Sbjct: 682 NDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNKKEGL-----IAXFDLG 825
Score = 69.3 bits (168), Expect(2) = 2e-36
Identities = 38/72 (52%), Positives = 46/72 (63%), Gaps = 1/72 (1%)
Frame = +3
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
IGIDLGTT SCV V +N +V I N +G RTTPS VAF + L+G K QA TNP
Sbjct: 255 IGIDLGTTNSCVSVMEGKNPKV--IENSEGARTTPSVVAFNQKGELLVGTPXKRQAVTNP 428
Query: 69 SNTIFAFKVIAG 80
+NT+F K + G
Sbjct: 429 TNTLFGTKRLIG 464
>BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chaperone hsp70 -
rice (fragment), partial (18%)
Length = 398
Score = 134 bits (338), Expect = 6e-32
Identities = 74/125 (59%), Positives = 85/125 (67%), Gaps = 22/125 (17%)
Frame = -3
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS V FT+++RLIGDAA
Sbjct: 369 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 196
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP NT+F FKV++G +KP I V+YKGE+K+F
Sbjct: 195 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVLSGPAEKPMIQVSYKGEDKQF 16
Query: 99 VAEEI 103
AEEI
Sbjct: 15 AAEEI 1
>TC207148 heat shock protein 70-related protein
Length = 2025
Score = 120 bits (302), Expect = 9e-28
Identities = 72/217 (33%), Positives = 114/217 (52%), Gaps = 9/217 (4%)
Frame = +2
Query: 209 AGDTHLGGQDFDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATI 268
A D LGG+DFD ++ HF +F+ +Y D+ +KA RLR ACEK K+ LS + +A +
Sbjct: 14 AFDRSLGGRDFDEVIFSHFAAKFKEEYHIDVYSKTKACSRLRAACEKLKKVLSANLEAPL 193
Query: 269 ELDAIYKGIDFCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSR 328
++ + G D ITR +FE++ L E+ L DA + + + V LVG SR
Sbjct: 194 NIECLMDGKDVKGFITREEFEKLASGLLERVSIPCRRALTDANLTEEKISSVELVGSGSR 373
Query: 329 IPKVRQLLQEVFKGKELCKSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGV 388
IP + LL +FK +E + +N E VA G +Q A+LS + V ++DV P S G+
Sbjct: 374 IPAISTLLTSLFK-REPSRQLNASECVARGCALQCAMLSPVYR-VREYEVKDVIPFSIGL 547
Query: 389 SELGNLMS-----VMIPRNTSIP----VTVEKTKTYY 416
S ++ V+ PR P +T +++ ++
Sbjct: 548 SSDEGPVAVRSNGVLFPRGQPFPSVKVITFQRSNLFH 658
>TC209896 similar to UP|O23508 (O23508) Growth regulator like protein,
partial (24%)
Length = 1185
Score = 112 bits (280), Expect = 3e-25
Identities = 69/219 (31%), Positives = 121/219 (54%), Gaps = 9/219 (4%)
Frame = +2
Query: 197 TVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKEFRNKYKE--DISENSKALRRLRTACE 254
+V N F+VK D LGGQ + +V++F +F + D+ + KA+ +L+ +
Sbjct: 14 SVSVNQFQVKDVRWDPELGGQHMELRLVEYFADQFNAQVGGGIDVRKFPKAMAKLKKQVK 193
Query: 255 KAKRTLSYDTDATIELDAIYKGIDFCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDK 314
+ K LS +T A I +++++ +DF S+ITR KFEE+ +++EK + V L ++ +
Sbjct: 194 RTKEILSANTAAPISVESLHDDVDFRSTITREKFEELCEDIWEKSLLPVKEVLENSGLSL 373
Query: 315 NSVDDVVLVGGSSRIPKVRQLLQEVFKGKELCKSINPDEAVAYGAVVQAALLSEGSKNVP 374
+ V L+GG++R+PK++ LQE + KEL + ++ DEA+ GA + AA LS+G K
Sbjct: 374 EQIYAVELIGGATRVPKLQAKLQEFLRRKELDRHLDADEAIVLGAALHAANLSDGIKLNR 553
Query: 375 NLVLRDVTPLSFGVSELG-------NLMSVMIPRNTSIP 406
L + D + F V G + +++PR +P
Sbjct: 554 KLGMIDGSLYGFVVELNGPDLLKDESSRQILVPRMKKVP 670
>TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (51%)
Length = 1228
Score = 110 bits (274), Expect = 2e-24
Identities = 63/152 (41%), Positives = 95/152 (62%)
Frame = +2
Query: 282 SITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFK 341
++TR+KFE + +L E+ CL DA + VD+V+LVGG +R+PKV++++ E+F
Sbjct: 41 TLTRSKFEALVNHLIERTKVPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF- 217
Query: 342 GKELCKSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPR 401
GK K +NPDEAVA GA +Q +L +V L+L DVTPLS G+ LG + + +I R
Sbjct: 218 GKSPSKGVNPDEAVAMGAAIQGGIL---RGDVKELLLLDVTPLSLGIETLGGIFTRLINR 388
Query: 402 NTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
NT+IP +K++ + D N+ V +KV Q
Sbjct: 389 NTTIP--TKKSQVFSTAAD--NQTQVGIKVLQ 472
>AW757289
Length = 402
Score = 104 bits (259), Expect = 9e-23
Identities = 51/97 (52%), Positives = 71/97 (72%), Gaps = 1/97 (1%)
Frame = +3
Query: 283 ITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKG 342
+TRA+FEE+N +LF K M V + DA + K+ +D++VLVGGS+RIPKV+QLL++ F G
Sbjct: 6 LTRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDG 185
Query: 343 KELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVL 378
KE K +NPDEAVAYGA VQ ++LS EG + ++
Sbjct: 186 KEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKGTIV 296
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,586,262
Number of Sequences: 63676
Number of extensions: 161013
Number of successful extensions: 863
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 813
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 813
length of query: 433
length of database: 12,639,632
effective HSP length: 100
effective length of query: 333
effective length of database: 6,272,032
effective search space: 2088586656
effective search space used: 2088586656
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144722.18


