

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.12 + phase: 0
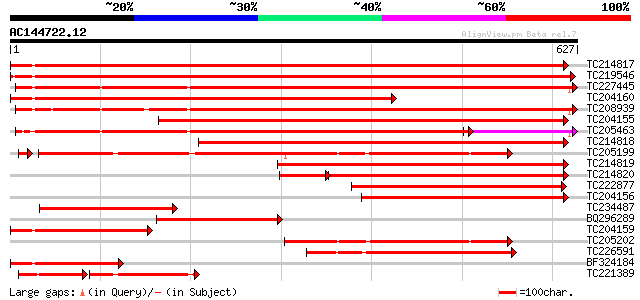
(627 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 810 0.0
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 780 0.0
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 629 0.0
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 609 e-174
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 608 e-174
TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, ... 580 e-166
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 531 e-151
TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous hea... 516 e-146
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 426 e-121
TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat sh... 380 e-105
TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight h... 284 2e-97
TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa p... 251 9e-67
TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 231 6e-61
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 226 3e-59
BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cyto... 220 2e-57
TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 207 9e-54
TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 202 3e-52
TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock ... 189 4e-48
BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chapero... 173 2e-43
TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa p... 104 4e-40
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 810 bits (2093), Expect = 0.0
Identities = 413/621 (66%), Positives = 502/621 (80%), Gaps = 3/621 (0%)
Frame = +2
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII N+QGNRTTPS+V F+ +RLIGDAA
Sbjct: 86 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 259
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR++SDS +++D++LWPFKVI G D P+IVV++K EEK F
Sbjct: 260 KNQVAMNPINTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIPGAADKPMIVVNYKGEEKQF 439
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIIN 180
AEEISSM+L KM+EIAE +L S VK+AV+TVPAYFNDSQR+ATKDAGVI+GLNV+RIIN
Sbjct: 440 AAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 619
Query: 181 EPTAAALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGED 240
EPTAAA+AYGL K+A V +N+ IFDLGGGTFDVS+LTI+ F+VKATAGDTHLGGED
Sbjct: 620 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 799
Query: 241 FDHRMVNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGID 300
FD+RMVNHFV+EFKRK+ KDISGN RALRRLRTACERAKRTLS + TI+ID++ EGID
Sbjct: 800 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGID 979
Query: 301 FCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQD 360
F S++TRA+FE+LNMDLF KCME VE+CL DA+MDK++VDDVVLVGGS+RIPKV+QLLQD
Sbjct: 980 FYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQLLQD 1159
Query: 361 FFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSV 419
FFNGK+LC SINPDEAVAYGAAVQAA+L GEG + V +L+L DVTPLSLG+ G VM+V
Sbjct: 1160FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1339
Query: 420 LIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP 478
LIPRN+ IP KK+QV+ T D+QPGV + V+EGER+ +NNLLG FEL +P APRG+P
Sbjct: 1340LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTRDNNLLGKFELSGIPPAPRGVP 1519
Query: 479 -IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKF 537
I VCF IDA+GILNVSAE++T+G K ITIT + GRLS E+IE+M+QEAE +K ED +
Sbjct: 1520QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEH 1699
Query: 538 KKKVKAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVF 597
KKKV+A N L++Y YNMR +KD + L DK K+ A+ + +D + E F
Sbjct: 1700KKKVEAKNALENYAYNMRNTVKDDKIGEKLDPADKKKIEDAIEQAIQWLDSNQLAEADEF 1879
Query: 598 VDFLKELEKIFESALNKINKG 618
D +KELE I + K+ +G
Sbjct: 1880EDKMKELESICNPIIAKMYQG 1942
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 780 bits (2014), Expect = 0.0
Identities = 403/628 (64%), Positives = 495/628 (78%), Gaps = 3/628 (0%)
Frame = +1
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EG AIGIDLGTTYSCV VWQ N+R EII N+QGNRTTPS+VAF+ +RLIGDAA
Sbjct: 1 MATK-EGKAIGIDLGTTYSCVGVWQ--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAA 171
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR++SDS ++ND++LWPFKV D P+IVV++K EEK F
Sbjct: 172 KNQVAMNPQNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCDKPMIVVNYKGEEKKF 351
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIIN 180
AEEISSM+L KM+E+AE FL VK+AV+TVPAYFNDSQR+ATKDAG ISGLNV+RIIN
Sbjct: 352 SAEEISSMVLVKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIIN 531
Query: 181 EPTAAALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGED 240
EPTAAA+AYGL K+A+ +N+ IFDLGGGTFDVS+LTI+ F+VKATAGDTHLGGED
Sbjct: 532 EPTAAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGED 711
Query: 241 FDHRMVNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGID 300
FD+RMVNHFV EFKRK+ KDISGN+RALRRLRTACERAKRTLS + TI+ID++ EGID
Sbjct: 712 FDNRMVNHFVSEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGID 891
Query: 301 FCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQD 360
F ++ITRA+FE++NMDLF KCME VE+CL DA++DK V +VVLVGGS+RIPKV QLLQD
Sbjct: 892 FYATITRARFEEMNMDLFRKCMEPVEKCLRDAKIDKSQVHEVVLVGGSTRIPKVHQLLQD 1071
Query: 361 FFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSV 419
FFNGK+LC SINPDEAVAYGAAVQAA+L G+G + V +L+L DVTPLSLG+ G VM+V
Sbjct: 1072FFNGKELCKSINPDEAVAYGAAVQAAILSGQGDEKVQDLLLLDVTPLSLGLETAGGVMTV 1251
Query: 420 LIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP 478
LIPRN+ IP KK+Q++ T D+QPGV + V+EGER+ +NNLLG FEL +P APRG+P
Sbjct: 1252LIPRNTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVP 1431
Query: 479 -IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKF 537
I VCF IDA+GILNVSAE++T+G K ITIT + GRLS EEIE+M+++AE +K ED +
Sbjct: 1432QINVCFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAERYKAEDEEV 1611
Query: 538 KKKVKAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVF 597
KKKV+A N L++Y YNMR +KD + L+ +K K+ A+ ++ + E F
Sbjct: 1612KKKVEAKNSLENYAYNMRNTIKDEKIGGKLSPDEKQKIEKAVEDAIQWLEGNQMAEVDEF 1791
Query: 598 VDFLKELEKIFESALNKINKGYSDEESD 625
D KELE I + K+ +G + D
Sbjct: 1792EDKQKELEGICNPIIAKMYQGAAGPGGD 1875
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 629 bits (1623), Expect = 0.0
Identities = 334/635 (52%), Positives = 446/635 (69%), Gaps = 14/635 (2%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII N+QGNR TPS+VAF+ +RLIG+AAKN AA
Sbjct: 209 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVAFTDSERLIGEAAKNLAAV 382
Query: 67 NPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKE-EEKHFVAEEI 125
NP T+FD KRLIGRK+ D ++ D++L P+K++ + P I V K+ E K F EEI
Sbjct: 383 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NKDGKPYIQVKIKDGETKVFSPEEI 559
Query: 126 SSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAA 185
S+MIL KMKE AE FL K+ DAV+TVPAYFND+QR+ATKDAGVI+GLNV RIINEPTAA
Sbjct: 560 SAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 739
Query: 186 ALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRM 245
A+AYGL K+ +NI +FDLGGGTFDVS+LTI N F+V AT GDTHLGGEDFD R+
Sbjct: 740 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 910
Query: 246 VNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCSSI 305
+ +F+K K+KH KDIS ++RAL +LR ERAKR LS + ++I+++ +G+DF +
Sbjct: 911 MEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEPL 1090
Query: 306 TRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGK 365
TRA+FE+LN DLF K M V++ + DA + K +D++VLVGGS+RIPKV+QLL+D+F+GK
Sbjct: 1091TRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1270
Query: 366 DLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRN 424
+ +NPDEAVAYGAAVQ ++L GEG + +++L DV PL+LGI G VM+ LIPRN
Sbjct: 1271EPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1450
Query: 425 SIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP-IQVC 482
++IP KK QV+ T D Q VS+ V+EGERS+ + LLG F+L +P APRG P I+V
Sbjct: 1451TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPRGTPQIEVT 1630
Query: 483 FAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVK 542
F +DA+GILNV AE++ +G + ITIT E GRLS EEI+RM++EAE F EED K K+++
Sbjct: 1631FEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKERID 1810
Query: 543 AMNVLDDYLYNMRKVMKD-GSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVFVDFL 601
A N L+ Y+YNM+ + D + L S +K K+ A+ + +DD + E + + L
Sbjct: 1811ARNSLETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSVEKEDYEEKL 1990
Query: 602 KELEKIFESALNKINK---------GYSDEESDSD 627
KE+E + ++ + + G S EE D
Sbjct: 1991KEVEAVCNPIISAVYQRSGGAPGGGGASGEEDKDD 2095
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 609 bits (1570), Expect = e-174
Identities = 308/428 (71%), Positives = 364/428 (84%), Gaps = 1/428 (0%)
Frame = +2
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII N+QGNRTTPS+VAF+ +RLIGDAA
Sbjct: 95 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 268
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR++SD+ ++ D++LWPFKVI G + P+IVV++K EEK F
Sbjct: 269 KNQVAMNPTNTVFDAKRLIGRRFSDASVQGDMKLWPFKVIPGPAEKPMIVVNYKGEEKQF 448
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIIN 180
AEEISSM+L KMKEIAE +L S +K+AV+TVPAYFNDSQR+ATKDAGVISGLNV+RIIN
Sbjct: 449 SAEEISSMVLMKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 628
Query: 181 EPTAAALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGED 240
EPTAAA+AYGL K+A +N+ IFDLGGGTFDVS+LTI+ F+VKATAGDTHLGGED
Sbjct: 629 EPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 808
Query: 241 FDHRMVNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGID 300
FD+RMVNHFV+EFKRK+ KDISGN+RALRRLRTACERAKRTLS + TI+ID++ EGID
Sbjct: 809 FDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGID 988
Query: 301 FCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQD 360
F ++ITRA+FE+LNMDLF KCME VE+CL DA+MDK TV DVVLVGGS+RIPKV+QLLQD
Sbjct: 989 FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 1168
Query: 361 FFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSV 419
FFNGK+LC SINPDEAVAYGAAVQAA+L GEG + V +L+L DVT LS + VM+V
Sbjct: 1169FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTTLSTALETARGVMTV 1348
Query: 420 LIPRNSII 427
L+ N+ I
Sbjct: 1349LMTSNTTI 1372
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 608 bits (1569), Expect = e-174
Identities = 329/634 (51%), Positives = 439/634 (68%), Gaps = 13/634 (2%)
Frame = +3
Query: 7 GPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII N+QGNR TPS+ +F+ +RLIG+AAKN AA
Sbjct: 171 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSW-SFTDSERLIGEAAKNLAAV 341
Query: 67 NPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKE-EEKHFVAEEI 125
NP +FD KRLIGRK+ D ++ D++L P+K++ + P I K+ E K F EEI
Sbjct: 342 NPERVIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NKDGKPYIQEKIKDGETKVFSPEEI 518
Query: 126 SSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAA 185
S+MIL KMKE AE FL K+ DAV AYFND+QR+ATKDAGVI+GLNV RIINEPTAA
Sbjct: 519 SAMILTKMKETAEAFLGKKINDAV----AYFNDAQRQATKDAGVIAGLNVARIINEPTAA 686
Query: 186 ALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRM 245
A+AYGL K+ +NI +FDLGGGTFDVS+LTI N F+V AT GDTHLGGEDFD R+
Sbjct: 687 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 857
Query: 246 VNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCSSI 305
+ +F+K +KH KDIS +SRAL +LR ERAKR LS + ++I+++ +G+DF +
Sbjct: 858 MEYFIKLINKKHKKDISKDSRALSKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEPL 1037
Query: 306 TRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGK 365
TRA+FE+LN DLF K M V++ + DA + K +D++VLVGGS+RIPKV+QLL+D+F+GK
Sbjct: 1038TRARFEELNNDLFRKTMGPVKKAMEDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1217
Query: 366 DLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRN 424
+ +NPDEAVAYGAAVQ ++L GEG + +++L DV PL+LGI G VM+ LIPRN
Sbjct: 1218EPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1397
Query: 425 SIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP-IQVC 482
++IP KK QV+ T D Q VS+ V+EGERS+ + LLG FEL +P APRG P I+V
Sbjct: 1398TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFELSGIPPAPRGTPQIEVT 1577
Query: 483 FAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVK 542
F +DA+GILNV AE++ +G + ITIT E GRLS EEIERM++EAE F EED K K+++
Sbjct: 1578FEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEEDKKVKERID 1757
Query: 543 AMNVLDDYLYNMRKVMKD-GSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVFVDFL 601
A N L+ Y+YNM+ + D + L S +K K+ A+ + +DD + E + + L
Sbjct: 1758ARNSLETYVYNMKNQVSDKDKLADKLESDEKEKVETAVKEALEWLDDNQSVEKEEYEEKL 1937
Query: 602 KELEKIFESALNKINK--------GYSDEESDSD 627
KE+E + ++ + + G S E+ D D
Sbjct: 1938KEVEAVCNPIISAVYQRSGGAPGGGASGEDDDED 2039
>TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, partial
(75%)
Length = 1640
Score = 580 bits (1495), Expect = e-166
Identities = 299/457 (65%), Positives = 363/457 (79%), Gaps = 3/457 (0%)
Frame = +2
Query: 165 KDAGVISGLNVIRIINEPTAAALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNA 224
KDAGVISGLNV+RIINEPTAAA+AYGL K+A +N+ IFDLGGGTFDVS+LTI+
Sbjct: 2 KDAGVISGLNVMRIINEPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGI 181
Query: 225 FDVKATAGDTHLGGEDFDHRMVNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSF 284
F+VKATAGDTHLGGEDFD+RMVNHFV+EFKRK+ KDISGN+RALRRLRTACERAKRTLS
Sbjct: 182 FEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSS 361
Query: 285 DTEATIDIDAISEGIDFCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVL 344
+ TI+ID++ EGIDF ++ITRA+FE+LNMDLF KCME VE+CL DA+MDK TV DVVL
Sbjct: 362 TAQTTIEIDSLYEGIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVL 541
Query: 345 VGGSSRIPKVKQLLQDFFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDV 403
VGGS+RIPKV+QLLQDFFNGK+LC SINPDEAVAYGAAVQAA+L GEG + V +L+L DV
Sbjct: 542 VGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDV 721
Query: 404 TPLSLGISIKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLL 463
TPLS G+ G VM+VLIPRN+ IP KK+QV+ T D+QPGV + VYEGER+ +NNLL
Sbjct: 722 TPLSTGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLL 901
Query: 464 GLFELK-VPLAPRGLP-IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIE 521
G FEL +P APRG+P I VCF IDA+GILNVSAE++T+G K ITIT + GRLS EEIE
Sbjct: 902 GKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIE 1081
Query: 522 RMIQEAENFKEEDLKFKKKVKAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMKLNAAMIK 581
+M+QEAE +K ED + KKKV+A N L++Y YNMR +KD + S L+S DK K+ A+ +
Sbjct: 1082KMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDDKIASKLSSDDKKKIEDAIEQ 1261
Query: 582 GKNLIDDKEQHETFVFVDFLKELEKIFESALNKINKG 618
+D + E F D +KELE I + K+ +G
Sbjct: 1262AIQWLDGNQLAEADEFEDKMKELESICNPIIAKMYQG 1372
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 531 bits (1369), Expect = e-151
Identities = 286/513 (55%), Positives = 371/513 (71%), Gaps = 6/513 (1%)
Frame = +3
Query: 7 GPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAAS 66
G IGIDL TTYSCV V + EII N+QGNR TPS+VAF+ +RLIG+AAK AA
Sbjct: 192 GTVIGIDL-TTYSCVGV----TDAVEIIANDQGNRITPSWVAFTDSERLIGEAAKIVAAV 356
Query: 67 NPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKE-EEKHFVAEEI 125
NP T+FD KRLIGRK+ D ++ D++L P+K++ + P I V K+ E K F EEI
Sbjct: 357 NPVRTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NKDGKPYIQVKIKDGETKVFSPEEI 533
Query: 126 SSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAA 185
S+MIL KMKE AE FL K+ DAV+TVPAYFND+QR+ATKDAGVI+GLNV RIINEPTAA
Sbjct: 534 SAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 713
Query: 186 ALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRM 245
A+AYGL K+ +NI +FDLGGGTFDVS+LTI N F+V AT GDTHLGGEDFD R+
Sbjct: 714 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 884
Query: 246 VNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCSSI 305
+ +F+K K+KH KDIS ++RAL +LR ERAKR LS + ++I+++ +G+DF +
Sbjct: 885 MEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEPL 1064
Query: 306 TRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGK 365
TRA+FE+LN DLF K M V++ + DA + K +D++VLVGGS+RIPKV+QLL+D+F+GK
Sbjct: 1065TRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1244
Query: 366 DLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRN 424
+ +NPDEAVAYGAAVQ ++L GEG + +++L DV PL+LGI G VM+ LIPRN
Sbjct: 1245EPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1424
Query: 425 SIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP-IQVC 482
++IP KK QV+ T D Q VS+ V+EGERS+ + LLG F+L +P APRG P I+V
Sbjct: 1425TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPRGTPQIEVT 1604
Query: 483 FAIDAD-GILNVSAEE-ETSGNKKDITITKENG 513
F +DA+ GILNV A + +G + IT K G
Sbjct: 1605FEVDAERGILNVKARRAKGTGKSEKITDYKRKG 1703
Score = 84.3 bits (207), Expect = 1e-16
Identities = 49/136 (36%), Positives = 75/136 (55%), Gaps = 10/136 (7%)
Frame = +1
Query: 502 NKKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVKAMNVLDDYLYNMRKVMKD- 560
N+K IT E GRLS EEIERM++EAE F EED K K+++ A N L+ Y+YNM+ + D
Sbjct: 1669 NQKRSPITNEKGRLSQEEIERMVREAEEFAEEDKKVKERIDARNSLETYVYNMKNQISDK 1848
Query: 561 GSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVFVDFLKELEKIFESALNKINK--- 617
+ L S +K K+ A+ + +DD + E + + LKE+E + ++ + +
Sbjct: 1849 DKLADKLESDEKEKIETAVKEALEWLDDNQSMEKEDYEEKLKEVEAVCNPIISAVYQRSG 2028
Query: 618 ------GYSDEESDSD 627
G S EE + D
Sbjct: 2029 GAPGGGGASGEEDEDD 2076
>TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (68%)
Length = 1554
Score = 516 bits (1328), Expect = e-146
Identities = 267/413 (64%), Positives = 324/413 (77%), Gaps = 3/413 (0%)
Frame = +2
Query: 209 GGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRMVNHFVKEFKRKHSKDISGNSRAL 268
GGGTFDVS+LTI+ F+VKATAGDTHLGGEDFD+RMVNHFV+EFKRK+ KDISGN RAL
Sbjct: 8 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRAL 187
Query: 269 RRLRTACERAKRTLSFDTEATIDIDAISEGIDFCSSITRAKFEQLNMDLFEKCMETVERC 328
RRLRTACERAKRTLS + TI+ID++ EGIDF S+ITRA+FE+LNMDLF KCME VE+C
Sbjct: 188 RRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYSTITRARFEELNMDLFRKCMEPVEKC 367
Query: 329 LTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGKDLCMSINPDEAVAYGAAVQAALL 388
L DA+MDK+TV DVVLVGGS+RIPKV+QLLQDFFNGK+LC SINPDEAVAYGAAVQAA+L
Sbjct: 368 LRDAKMDKRTVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAIL 547
Query: 389 -GEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSV 447
GEG + V +L+L DVTPLSLG+ G VM+VLIPRN+ IP KK+QV+ T D+QPGV +
Sbjct: 548 SGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLI 727
Query: 448 DVYEGERSVASENNLLGLFELK-VPLAPRGLP-IQVCFAIDADGILNVSAEEETSGNKKD 505
VYEGER+ +NNLLG FEL +P APRG+P I VCF IDA+GILNVSAE++T+G K
Sbjct: 728 QVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNK 907
Query: 506 ITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVKAMNVLDDYLYNMRKVMKDGSVTS 565
ITIT + GRLS EEIE+M+QEAE +K ED + KKKV+A N L++Y YNMR +KD +
Sbjct: 908 ITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKKKVEAKNALENYSYNMRNTIKDEKIGG 1087
Query: 566 MLTSTDKMKLNAAMIKGKNLIDDKEQHETFVFVDFLKELEKIFESALNKINKG 618
L DK K+ A+ + +D + E F D +KELE I + K+ +G
Sbjct: 1088KLDPADKKKIEDAIEQAIQWLDSNQLGEADEFEDKMKELESICNPIIAKMYQG 1246
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 426 bits (1096), Expect(2) = e-121
Identities = 239/532 (44%), Positives = 347/532 (64%), Gaps = 7/532 (1%)
Frame = +3
Query: 32 EIIHNEQGNRTTPSFVAFSGD-QRLIGDAAKNQAASNPANTVFDAKRLIGRKYSDSVIKN 90
++I N +G RTTPS VAF+ + L+G AK QA +NP NT+F KRLIGR++ DS +
Sbjct: 186 KVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTKRLIGRRFDDSQTQK 365
Query: 91 DLQLWPFKVIAGTEDNPVIVVHFKEEEKHFVAEEISSMILAKMKEIAEKFLESKVKDAVI 150
++++ P+K++ + + + ++ + ++ + +L KMKE AE +L V AVI
Sbjct: 366 EMKMVPYKIVKAPNGDAWVEANGQQ----YSPSQVGAFVLTKMKETAESYLGKSVSKAVI 533
Query: 151 TVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAAALAYGLQKRANCVENRNIFIFDLGG 210
TVPAYFND+QR+ATKDAG I+GL+V RIINEPTAAAL+YG+ + + +FDLGG
Sbjct: 534 TVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNNKEGLIA-----VFDLGG 698
Query: 211 GTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRMVNHFVKEFKRKHSKDISGNSRALRR 270
GTFDVS+L I N F+VKAT GDT LGGEDFD+ +++ V EFKR + D+S + AL+R
Sbjct: 699 GTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRTENIDLSKDKLALQR 878
Query: 271 LRTACERAKRTLSFDTEATIDIDAISEGIDFCS----SITRAKFEQLNMDLFEKCMETVE 326
LR A E+AK LS ++ I++ I+ ++TR+KFE L L E+ +
Sbjct: 879 LREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNHLIERTKAPCK 1058
Query: 327 RCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGKDLCMSINPDEAVAYGAAVQAA 386
CL DA + K VD+V+LVGG +R+PKV++++ F GK +NPDEAVA GAA+Q
Sbjct: 1059SCLKDANVSIKEVDEVLLVGGMTRVPKVQEVVSAIF-GKSPSKGVNPDEAVAMGAAIQGG 1235
Query: 387 LLGEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQPGVS 446
+L +K L+L DVTPLSLGI G + + LI RN+ IP KK QV+ T D+Q V
Sbjct: 1236ILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVG 1406
Query: 447 VDVYEGERSVASENNLLGLFEL-KVPLAPRGLP-IQVCFAIDADGILNVSAEEETSGNKK 504
+ V +GER +A +N LG FEL +P APRG+P I+V F IDA+GI+ VSA+++++G ++
Sbjct: 1407IKVLQGEREMAVDNKSLGEFELVGIPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQ 1586
Query: 505 DITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVKAMNVLDDYLYNMRK 556
ITI + +G LS +EI++M++EAE ++D + K + N D +Y++ K
Sbjct: 1587QITI-RSSGGLSEDEIDKMVKEAELHAQKDQERKALIDIRNSADTSIYSIEK 1739
Score = 28.1 bits (61), Expect(2) = e-121
Identities = 12/16 (75%), Positives = 14/16 (87%)
Frame = +1
Query: 10 IGIDLGTTYSCVAVWQ 25
IGIDLGTT SCV+V +
Sbjct: 49 IGIDLGTTNSCVSVME 96
>TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat shock cognate
protein 70, partial (54%)
Length = 1266
Score = 380 bits (975), Expect = e-105
Identities = 197/325 (60%), Positives = 248/325 (75%), Gaps = 3/325 (0%)
Frame = +1
Query: 297 EGIDFCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQ 356
EGIDF S++TRA+FE+LNMDLF KCME VE+CL DA+MDK++VDDVVLVGGS+RIPKV+Q
Sbjct: 4 EGIDFYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQ 183
Query: 357 LLQDFFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGD 415
LLQDFFNGK+LC SINPDEAVAYGAAVQAA+L GEG + V +L+L DVTPLSLG+ G
Sbjct: 184 LLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGG 363
Query: 416 VMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAP 474
VM+VLIPRN+ IP KK+QV+ T D+QPGV + V+EGER+ +NNLLG FEL +P AP
Sbjct: 364 VMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELSGIPPAP 543
Query: 475 RGLP-IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEE 533
RG+P I VCF IDA+GILNVSAE++T+G K ITIT + GRLS E+IE+M+QEAE +K E
Sbjct: 544 RGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSE 723
Query: 534 DLKFKKKVKAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHE 593
D + KKKV+A N L++Y YNMR +KD + L TDK K+ A+ + +D + E
Sbjct: 724 DEEHKKKVEAKNALENYAYNMRNTVKDDKIGEKLDPTDKKKIEDAIEQAIQWLDSNQLAE 903
Query: 594 TFVFVDFLKELEKIFESALNKINKG 618
F D +KELE I + K+ +G
Sbjct: 904 ADEFEDKMKELESICNPIIAKMYQG 978
>TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight heat shock
protein, partial (54%)
Length = 1272
Score = 284 bits (727), Expect(2) = 2e-97
Identities = 152/269 (56%), Positives = 194/269 (71%), Gaps = 3/269 (1%)
Frame = +2
Query: 353 KVKQLLQDFFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGIS 411
K +QL+QDF+NGK+LC SINPDEAVAYGAAVQAA+L GEG + V +L+L DVTPLSLG+
Sbjct: 164 K*QQLVQDFYNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLE 343
Query: 412 IKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-V 470
G VM+VLIPRN+ IP KK+QV+ T D+QPGV + VYEGER+ +NNLLG FEL +
Sbjct: 344 TAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGI 523
Query: 471 PLAPRGLP-IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAEN 529
P APRG+P I VCF IDA+GILNVSAE++T+G K ITIT + GRLS +EIE+M+QEAE
Sbjct: 524 PPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQEAEK 703
Query: 530 FKEEDLKFKKKVKAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMKLNAAMIKGKNLIDDK 589
+K ED + KKKV A N L++Y YNMR +KD + S L+ DK K+ A+ +D
Sbjct: 704 YKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIASKLSDDDKKKIEDAIESAIQWLDGN 883
Query: 590 EQHETFVFVDFLKELEKIFESALNKINKG 618
+ E F D +KELE I + K+ +G
Sbjct: 884 QLAEADEFEDKMKELESICNPIIAKMYQG 970
Score = 90.9 bits (224), Expect(2) = 2e-97
Identities = 42/56 (75%), Positives = 50/56 (89%)
Frame = +1
Query: 299 IDFCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKV 354
+DF ++ITRA+FE+LNMDLF KCME VE+CL DA+MDK TV DVVLVGGS+RIPKV
Sbjct: 1 VDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKV 168
>TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa protein
(Fragment), partial (19%)
Length = 809
Score = 251 bits (640), Expect = 9e-67
Identities = 133/241 (55%), Positives = 180/241 (74%), Gaps = 4/241 (1%)
Frame = +1
Query: 379 YGAAVQAALLGEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRNSIIPVKKKQVYHTC 438
YGAAVQAALL +GI +VPNLVL D+TPLSLG+S++GD+MSV+IPRN+ IPV++ + Y T
Sbjct: 1 YGAAVQAALLSKGIVNVPNLVLLDITPLSLGVSVQGDLMSVVIPRNTTIPVRRTKTYVTT 180
Query: 439 DDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLPIQVCFAIDADGILNVSAEE 497
+D+Q V ++VYEGER+ AS+NNLLG F L +P APRG P+ F ID +GIL+VSAEE
Sbjct: 181 EDNQSAVMIEVYEGERTRASDNNLLGFFTLSGIPPAPRGHPLYETFDIDENGILSVSAEE 360
Query: 498 ETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVKAMNVLDDYLYNMRKV 557
E++GNK +ITIT E RLST+EI+RMIQEAE +K ED KF +K KAMN LD Y+Y ++
Sbjct: 361 ESTGNKNEITITNEKERLSTKEIKRMIQEAEYYKAEDKKFLRKAKAMNDLDYYVYKIKNA 540
Query: 558 MKDGSVTSMLTSTDKMKLNAAMIKGKNLIDD-KEQHETFVFVDFLKELEKIFE--SALNK 614
+K ++S L S +K +++A+ + +L++D +Q + VF D LKELE I E A+ K
Sbjct: 541 LKKKDISSKLCSKEKENVSSAIARATDLLEDNNQQDDIVVFEDNLKELESIIERMKAMGK 720
Query: 615 I 615
I
Sbjct: 721 I 723
>TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (40%)
Length = 965
Score = 231 bits (590), Expect = 6e-61
Identities = 123/231 (53%), Positives = 162/231 (69%), Gaps = 2/231 (0%)
Frame = +2
Query: 390 EGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSVDV 449
EG + V +L+L DVTPLSLG+ G VM+VLIPRN+ IP KK+QV+ T D+QPGV + V
Sbjct: 2 EGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQV 181
Query: 450 YEGERSVASENNLLGLFELK-VPLAPRGLP-IQVCFAIDADGILNVSAEEETSGNKKDIT 507
YEGER+ +NNLLG FEL +P APRG+P I VCF IDA+GILNVSAE++T+G K IT
Sbjct: 182 YEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKIT 361
Query: 508 ITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVKAMNVLDDYLYNMRKVMKDGSVTSML 567
IT + GRLS EEIE+M+QEAE +K ED + KKKV+A N L++Y YNMR +KD + S L
Sbjct: 362 ITNDKGRLSKEEIEKMVQEAEKYKAEDEEHKKKVEAKNTLENYAYNMRNTIKDDKIASKL 541
Query: 568 TSTDKMKLNAAMIKGKNLIDDKEQHETFVFVDFLKELEKIFESALNKINKG 618
++ DK K+ A+ + +D + E F D +KELE I + K+ +G
Sbjct: 542 SADDKKKIEDAIEQAIQWLDGNQLAEADEFEDKMKELESICNPIIAKMYQG 694
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 226 bits (575), Expect = 3e-59
Identities = 109/152 (71%), Positives = 132/152 (86%)
Frame = +1
Query: 34 IHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQ 93
IHN+QGN TTPS VAF+ QRLIG+AAKNQAA+NP NTVFDAKRLIGRK+SD VI+ D
Sbjct: 1 IHNDQGNNTTPSCVAFTDQQRLIGEAAKNQAATNPENTVFDAKRLIGRKFSDPVIQKDKM 180
Query: 94 LWPFKVIAGTEDNPVIVVHFKEEEKHFVAEEISSMILAKMKEIAEKFLESKVKDAVITVP 153
LWPFKV+AG D P+I +++K +EKH +AEE+SSM+L KM+EIAE +LE+ V++AV+TVP
Sbjct: 181 LWPFKVVAGINDKPMISLNYKGQEKHLLAEEVSSMVLIKMREIAEAYLETPVENAVVTVP 360
Query: 154 AYFNDSQRKATKDAGVISGLNVIRIINEPTAA 185
AYFNDSQRKAT DAG I+GLNV+RIINEPTAA
Sbjct: 361 AYFNDSQRKATIDAGAIAGLNVMRIINEPTAA 456
>BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cytosolic
[imported] - spinach, partial (21%)
Length = 421
Score = 220 bits (560), Expect = 2e-57
Identities = 106/139 (76%), Positives = 123/139 (88%)
Frame = +1
Query: 163 ATKDAGVISGLNVIRIINEPTAAALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKN 222
ATKDAGVI+GLNV+RIINEPTAAA+AYGL K+A V +N+ IFDLGGGTFDVS+LTI+
Sbjct: 1 ATKDAGVIAGLNVMRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE 180
Query: 223 NAFDVKATAGDTHLGGEDFDHRMVNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTL 282
F+VKATAGDTHLGGEDFD+RMVNHFV+EFKRKH KDI+GN RALRRLRTACERAKRTL
Sbjct: 181 GIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKHKKDINGNPRALRRLRTACERAKRTL 360
Query: 283 SFDTEATIDIDAISEGIDF 301
S + TI+ID++ EG+DF
Sbjct: 361 SSTAQTTIEIDSLYEGVDF 417
>TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (22%)
Length = 769
Score = 207 bits (528), Expect = 9e-54
Identities = 106/159 (66%), Positives = 128/159 (79%), Gaps = 1/159 (0%)
Frame = +2
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII N+QGNRTTPS+VAF+ +RLIGDAA
Sbjct: 80 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 253
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR++SD+ +++D++LWPFKVI G D P+IVV++K +EK F
Sbjct: 254 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKVIPGPADKPMIVVNYKGDEKQF 433
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVP-AYFND 158
AEEISSM+L KMKEIAE +L S +K+A YFND
Sbjct: 434 SAEEISSMVLIKMKEIAEAYLGSTIKNASCHPSLLYFND 550
>TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (51%)
Length = 1228
Score = 202 bits (515), Expect = 3e-52
Identities = 114/255 (44%), Positives = 170/255 (65%), Gaps = 2/255 (0%)
Frame = +2
Query: 304 SITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFN 363
++TR+KFE L L E+ + CL DA + K VD+V+LVGG +R+PKV++++ + F
Sbjct: 41 TLTRSKFEALVNHLIERTKVPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF- 217
Query: 364 GKDLCMSINPDEAVAYGAAVQAALLGEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPR 423
GK +NPDEAVA GAA+Q +L +K L+L DVTPLSLGI G + + LI R
Sbjct: 218 GKSPSKGVNPDEAVAMGAAIQGGILRGDVKE---LLLLDVTPLSLGIETLGGIFTRLINR 388
Query: 424 NSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFEL-KVPLAPRGLP-IQV 481
N+ IP KK QV+ T D+Q V + V +GER +A++N +LG F+L +P APRGLP I+V
Sbjct: 389 NTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMAADNKMLGEFDLVGIPPAPRGLPQIEV 568
Query: 482 CFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKV 541
F IDA+GI+ VSA+++++G ++ ITI + +G LS +EIE+M++EAE ++D + K +
Sbjct: 569 TFDIDANGIVTVSAKDKSTGKEQQITI-RSSGGLSEDEIEKMVKEAELHAQKDQERKALI 745
Query: 542 KAMNVLDDYLYNMRK 556
N D +Y++ K
Sbjct: 746 DIRNSADTTIYSIEK 790
>TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock protein
hsp70. {Pisum sativum;} , partial (45%)
Length = 1338
Score = 189 bits (479), Expect = 4e-48
Identities = 106/234 (45%), Positives = 155/234 (65%), Gaps = 2/234 (0%)
Frame = +3
Query: 329 LTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGKDLCMSINPDEAVAYGAAVQAALL 388
L DA++ K +D+V+LVGGS+RIP V++L++ GKD +++NPDE VA GAAVQA +L
Sbjct: 3 LRDAKLSFKDLDEVILVGGSTRIPAVQELVKKL-TGKDPNVTVNPDEVVALGAAVQAGVL 179
Query: 389 GEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSVD 448
V ++VL DVTPLSLG+ G VM+ +IPRN+ +P K +V+ T D Q V ++
Sbjct: 180 AG---DVSDIVLLDVTPLSLGLETLGGVMTKIIPRNTTLPTSKSEVFSTAADGQTSVEIN 350
Query: 449 VYEGERSVASENNLLGLFELK-VPLAPRGLP-IQVCFAIDADGILNVSAEEETSGNKKDI 506
V +GER +N LG F L +P APRG+P I+V F IDA+GIL+V+A ++ +G K+DI
Sbjct: 351 VLQGEREFVRDNKSLGSFRLDGIPPAPRGVPQIEVKFDIDANGILSVTAIDKGTGKKQDI 530
Query: 507 TITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVKAMNVLDDYLYNMRKVMKD 560
TIT + L ++E+ERM+ EAE F +ED + + + N D +Y K +K+
Sbjct: 531 TITGAS-TLPSDEVERMVNEAEKFSKEDKEKRDAIDTKNQADSVVYQTEKQLKE 689
>BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chaperone hsp70 -
rice (fragment), partial (18%)
Length = 398
Score = 173 bits (438), Expect = 2e-43
Identities = 83/125 (66%), Positives = 103/125 (82%)
Frame = -3
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII N+QGNRTTPS+V F+ +RLIGDAA
Sbjct: 369 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 196
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR++SD+ +++D++LWPFKV++G + P+I V +K E+K F
Sbjct: 195 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVLSGPAEKPMIQVSYKGEDKQF 16
Query: 121 VAEEI 125
AEEI
Sbjct: 15 AAEEI 1
>TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (36%)
Length = 827
Score = 104 bits (260), Expect(2) = 4e-40
Identities = 56/121 (46%), Positives = 79/121 (65%)
Frame = +1
Query: 89 KNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHFVAEEISSMILAKMKEIAEKFLESKVKDA 148
K ++++ PFK++ N + + ++ + +I + +L KMKE AE +L + A
Sbjct: 490 KREMKMVPFKIVKAPNGNAWVETNGQQ----YSPSQIGAFVLTKMKETAEAYLGKSISKA 657
Query: 149 VITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAAALAYGLQKRANCVENRNIFIFDL 208
VITVPAYFND+QR+ATKDAG I+GL+V RIINEPTAAAL+YG+ K+ I FDL
Sbjct: 658 VITVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNKKEGL-----IAXFDL 822
Query: 209 G 209
G
Sbjct: 823 G 825
Score = 79.3 bits (194), Expect(2) = 4e-40
Identities = 41/78 (52%), Positives = 55/78 (69%), Gaps = 1/78 (1%)
Frame = +3
Query: 10 IGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGD-QRLIGDAAKNQAASNP 68
IGIDLGTT SCV+V + +N + +I N +G RTTPS VAF+ + L+G K QA +NP
Sbjct: 255 IGIDLGTTNSCVSVMEGKNPK--VIENSEGARTTPSVVAFNQKGELLVGTPXKRQAVTNP 428
Query: 69 ANTVFDAKRLIGRKYSDS 86
NT+F KRLIGR++ D+
Sbjct: 429 TNTLFGTKRLIGRRFDDA 482
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,978,063
Number of Sequences: 63676
Number of extensions: 233875
Number of successful extensions: 1130
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 1059
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1064
length of query: 627
length of database: 12,639,632
effective HSP length: 103
effective length of query: 524
effective length of database: 6,081,004
effective search space: 3186446096
effective search space used: 3186446096
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144722.12


