

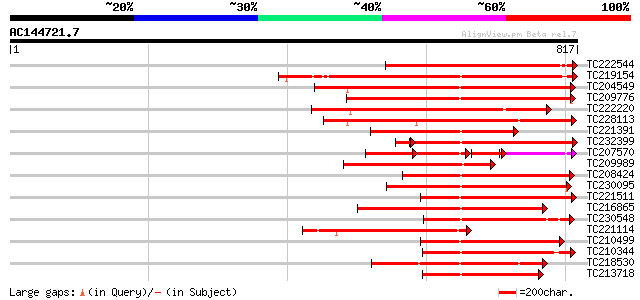
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144721.7 + phase: 0 /pseudo
(817 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precu... 429 e-120
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 390 e-108
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 375 e-104
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 375 e-104
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 345 4e-95
TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 340 1e-93
TC221391 similar to UP|O81906 (O81906) Serine/threonine kinase-l... 313 3e-85
TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial... 286 3e-77
TC207570 weakly similar to UP|O65470 (O65470) Serine/threonine k... 99 6e-71
TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 260 2e-69
TC208424 weakly similar to UP|O65466 (O65466) Serine/threonine k... 256 2e-68
TC230095 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like prot... 254 8e-68
TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, ... 251 7e-67
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 249 3e-66
TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-l... 235 7e-62
TC221114 similar to UP|Q8RZ15 (Q8RZ15) Receptor kinase-like prot... 234 1e-61
TC210499 weakly similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine k... 231 7e-61
TC210344 similar to UP|Q9AVE0 (Q9AVE0) SRKb, partial (21%) 231 1e-60
TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partia... 230 2e-60
TC213718 similar to UP|Q70I29 (Q70I29) S-receptor kinase-like pr... 227 1e-59
>TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precursor ,
partial (27%)
Length = 861
Score = 429 bits (1104), Expect = e-120
Identities = 217/276 (78%), Positives = 244/276 (87%)
Frame = +3
Query: 542 EVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNII 601
EVVVISKLQHRNLVRLLGCC+ER EQ+LVYEFMPNKSLD+FLFDPLQ+K LDW+KR NII
Sbjct: 3 EVVVISKLQHRNLVRLLGCCIERDEQMLVYEFMPNKSLDSFLFDPLQRKILDWKKRFNII 182
Query: 602 EGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRV 661
EGIARGI+YLHRDSRLRIIHRDLKASNILLD +M PKISDFGLARIV+ G+DDEANTKRV
Sbjct: 183 EGIARGILYLHRDSRLRIIHRDLKASNILLDDEMHPKISDFGLARIVRSGDDDEANTKRV 362
Query: 662 VGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWL 721
VGTYGYMPPEYAMEG+FSEKSDVY VLLLEIVSGRRN+SF ++E +LSL +AWKLW
Sbjct: 363 VGTYGYMPPEYAMEGIFSEKSDVYXXXVLLLEIVSGRRNTSFYNNEQSLSLXXYAWKLWN 542
Query: 722 EENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPG 781
E NI S+ID E+ D FE S+LRCIHIGLLCVQEL ++RP ISTVV ML+SEITHLPPP
Sbjct: 543 EGNIKSIIDLEIQDPMFEKSILRCIHIGLLCVQELTKERPTISTVVXMLISEITHLPPPR 722
Query: 782 RVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
+ FV KQ+ +S +ESSQKS Q NSNNNVT+SE+QG
Sbjct: 723 QXXFVQKQNCQS-SESSQKS-QFNSNNNVTISEIQG 824
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 390 bits (1002), Expect = e-108
Identities = 212/439 (48%), Positives = 298/439 (67%), Gaps = 9/439 (2%)
Frame = +1
Query: 388 SCLAYA-YDPS---IFCMYWTGELIDLQKFP--NGGVDLFVRVP-AELVAVKKEKGHNKS 440
SC+AY Y+ S C+ W G+L+D++ + G L +R+P +EL ++K +K +K
Sbjct: 1 SCMAYTNYNISGAGSGCVMWFGDLLDIKLYSVAESGRRLHIRLPPSELESIKSKKS-SKI 177
Query: 441 FLIIVIAGVIGALILVICAYLLWRKCSARHKAREH--QQMKLDELPLYDFEKLETATNCF 498
+ +A +G ++L IC ++ R + + K ++ +Q++ ++PL+D + AT+ F
Sbjct: 178 IIGTSVAAPLG-VVLAIC-FIYRRNIADKSKTKKSIDRQLQDVDVPLFDMLTITAATDNF 351
Query: 499 HFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLL 558
NN +G+GGFGPVYKG + GQEIAVKRLS SGQGI EF+ EV +I+KLQHRNLV+LL
Sbjct: 352 LLNNKIGEGGFGPVYKGKLVGGQEIAVKRLSSLSGQGITEFITEVKLIAKLQHRNLVKLL 531
Query: 559 GCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLR 618
GCC++ E++LVYE++ N SL++F+FD ++ K LDW +R NII GIARG++YLH+DSRLR
Sbjct: 532 GCCIKGQEKLLVYEYVVNGSLNSFIFDQIKSKLLDWPRRFNIILGIARGLLYLHQDSRLR 711
Query: 619 IIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLF 678
IIHRDLKASN+LLD + PKISDFG+AR G+ E NT RVVGTYGYM PEYA +G F
Sbjct: 712 IIHRDLKASNVLLDEKLNPKISDFGMARAFG-GDQTEGNTNRVVGTYGYMAPEYAFDGNF 888
Query: 679 SEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACF 738
S KSDV+SFG+LLLEIV G +N S H TL+LVG+AW LW E+N + LID + D+C
Sbjct: 889 SIKSDVFSFGILLLEIVCGIKNKSLCHENQTLNLVGYAWALWKEQNALQLIDSGIKDSCV 1068
Query: 739 ESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESS 798
+LRCIH+ LLCVQ+ P DRP +++V+ ML SE+ + P F + + +
Sbjct: 1069IPEVLRCIHVSLLCVQQYPEDRPTMTSVIQMLGSEMDMVEPKEPGFFPRRILKEGNLK-- 1242
Query: 799 QKSHQSNSNNNVTLSEVQG 817
+ SN+ +T+S G
Sbjct: 1243----EMTSNDELTISLFSG 1287
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 375 bits (964), Expect = e-104
Identities = 201/384 (52%), Positives = 262/384 (67%), Gaps = 8/384 (2%)
Frame = +3
Query: 440 SFLIIVIAGVIGALILVICAYLLWRKCSARHKAREHQQMKLDELPL-----YDFEKLETA 494
+ + IV+ + LI ++ L R+ + + + ++P +DF +E A
Sbjct: 639 TIVAIVVPITVAVLIFIVGICFLSRRARKKQQGSVKEGKTAYDIPTVDSLQFDFSTIEAA 818
Query: 495 TNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNL 554
TN F +N LG+GGFG VYKG + GQ +AVKRLSK+SGQG EEF NEVVV++KLQHRNL
Sbjct: 819 TNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRLSKSSGQGGEEFKNEVVVVAKLQHRNL 998
Query: 555 VRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRD 614
VRLLG C++ E+ILVYE++PNKSLD LFDP +++ LDW +R II GIARGI YLH D
Sbjct: 999 VRLLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQRELDWGRRYKIIGGIARGIQYLHED 1178
Query: 615 SRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDD-EANTKRVVGTYGYMPPEYA 673
SRLRIIHRDLKASNILLD DM PKISDFG+ARI FG D + NT R+VGTYGYM PEYA
Sbjct: 1179 SRLRIIHRDLKASNILLDGDMNPKISDFGMARI--FGVDQTQGNTSRIVGTYGYMAPEYA 1352
Query: 674 MEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEV 733
M G FS KSDVYSFGVLL+EI+SG++NSSF + L+ +AW+LW + + L+DP +
Sbjct: 1353 MHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTDGAEDLLSYAWQLWKDGTPLELMDPIL 1532
Query: 734 WDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAFVHKQSSKS 793
++ ++ ++R IHIGLLCVQE P DRP ++T+VLML S LP P + AF +
Sbjct: 1533 RESYNQNEVIRSIHIGLLCVQEDPADRPTMATIVLMLDSNTVTLPTPTQPAFFVHSGTDP 1712
Query: 794 TTESSQKSHQS--NSNNNVTLSEV 815
QS S N++++SE+
Sbjct: 1713 NMPKELPFDQSIPMSVNDMSISEM 1784
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 375 bits (963), Expect = e-104
Identities = 196/337 (58%), Positives = 245/337 (72%), Gaps = 7/337 (2%)
Frame = +2
Query: 486 YDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVV 545
+DF +E AT F N LG+GGFG VYKG++ GQE+AVKRLSK SGQG EEF NEV +
Sbjct: 44 FDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNEVEI 223
Query: 546 ISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIA 605
++KLQHRNLVRLLG C+E E+ILVYEF+ NKSLD LFDP ++K+LDW +R I+EGIA
Sbjct: 224 VAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWTRRYKIVEGIA 403
Query: 606 RGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDD-EANTKRVVGT 664
RGI YLH DSRL+IIHRDLKASN+LLD DM PKISDFG+ARI FG D +ANT R+VGT
Sbjct: 404 RGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARI--FGVDQTQANTNRIVGT 577
Query: 665 YGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEEN 724
YGYM PEYAM G +S KSDVYSFGVL+LEI+SG+RNSSF + L+ +AWKLW +E
Sbjct: 578 YGYMSPEYAMHGEYSAKSDVYSFGVLILEIISGKRNSSFYETDVAEDLLSYAWKLWKDEA 757
Query: 725 IISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVA 784
+ L+D + ++ + ++RCIHIGLLCVQE P DRP +++VVLML S L P + A
Sbjct: 758 PLELMDQSLRESYTRNEVIRCIHIGLLCVQEDPIDRPTMASVVLMLDSYSVTLQVPNQPA 937
Query: 785 FVHKQSSKSTTESSQKSHQSNSN------NNVTLSEV 815
F ++ K QS +N N++++SEV
Sbjct: 938 FYINSRTEPNMPKGLKIDQSTTNSTSKSVNDMSVSEV 1048
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 345 bits (886), Expect = 4e-95
Identities = 180/355 (50%), Positives = 248/355 (69%), Gaps = 10/355 (2%)
Frame = +3
Query: 436 GHNKSF--LIIVIAGVIGALILVICA-YLLWRKCSARHKAREHQQMKLDELPLYDFE--- 489
G+ KS +++++ V+ ++IL++C Y + ++ ++ + + L +
Sbjct: 18 GNKKSVSRVVLIVVPVVVSIILLLCVCYFILKRSRKKYNTLLRENFGEESATLESLQFGL 197
Query: 490 -KLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISK 548
+E ATN F + +G+GGFG VYKGV+ DG+EIAVK+LSK+SGQG EF NE+++I+K
Sbjct: 198 VTIEAATNKFSYEKRIGEGGFGVVYKGVLPDGREIAVKKLSKSSGQGANEFKNEILLIAK 377
Query: 549 LQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGI 608
LQHRNLV LLG C+E E++L+YEF+ NKSLD FLFD + K L+W +R IIEGIA+GI
Sbjct: 378 LQHRNLVTLLGFCLEEHEKMLIYEFVSNKSLDYFLFDSHRSKQLNWSERYKIIEGIAQGI 557
Query: 609 MYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYM 668
YLH SRL++IHRDLK SN+LLDS+M PKISDFG+ARIV + + T R+VGTYGYM
Sbjct: 558 SYLHEHSRLKVIHRDLKPSNVLLDSNMNPKISDFGMARIVAI-DQLQGKTNRIVGTYGYM 734
Query: 669 PPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRN--SSFSHHEDTLSLVGFAWKLWLEENII 726
PEYAM G FSEKSDV+SFGV++LEI+S +RN S FS H+D LS + W+ W++E +
Sbjct: 735 SPEYAMHGQFSEKSDVFSFGVIVLEIISAKRNTRSVFSDHDDLLS---YTWEQWMDEAPL 905
Query: 727 SLIDPEV-WDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPP 780
++ D + + C S +++CI IGLLCVQE P DRP I+ V+ L S IT LP P
Sbjct: 906 NIFDQSIEAEFCDHSEVVKCIQIGLLCVQEKPDDRPKITQVISYLNSSITELPLP 1070
>TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (53%)
Length = 1492
Score = 340 bits (873), Expect = 1e-93
Identities = 187/407 (45%), Positives = 252/407 (60%), Gaps = 43/407 (10%)
Frame = +1
Query: 453 LILVICAYLLWRKCSARHKAREHQQMKLDELPL-----YDFEKLETATNCFHFNNMLGKG 507
++ +IC + R+ AR + Q DE+ + ++F+ + AT F +N LG+G
Sbjct: 4 VVPLICLCIYSRRSKARKSSLVKQHEDDDEIEIAQSLQFNFDTIRVATEDFSDSNKLGQG 183
Query: 508 GFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQ 567
GFG VY+G + DGQ IAVKRLS+ S QG EF NEV++++KLQHRNLVRLLG C+E E+
Sbjct: 184 GFGAVYRGRLSDGQMIAVKRLSRESSQGDTEFKNEVLLVAKLQHRNLVRLLGFCLEGKER 363
Query: 568 ILVYEFMPNKSLDAFLF------------------------------------DPLQKKN 591
+L+YE++PNKSLD F+F DP +K
Sbjct: 364 LLIYEYVPNKSLDYFIFGRS**VHN*TVALYFTSGSGQRLNIHIPLKLMAVYADPTKKAQ 543
Query: 592 LDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFG 651
L+W R II G+ARG++YLH DS LRIIHRDLKASNILL+ +M PKI+DFG+AR+V
Sbjct: 544 LNWEMRYKIITGVARGLLYLHEDSHLRIIHRDLKASNILLNEEMNPKIADFGMARLVLM- 720
Query: 652 EDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLS 711
+ +ANT R+VGTYGYM PEYAM G FS KSDV+SFGVL+LEI+SG +NS H E+
Sbjct: 721 DQTQANTNRIVGTYGYMAPEYAMHGQFSMKSDVFSFGVLVLEIISGHKNSGIRHGENVED 900
Query: 712 LVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLV 771
L+ FAW+ W E + ++DP + + + MLRCIHIGLLCVQE DRP ++T++LML
Sbjct: 901 LLSFAWRNWREGTAVKIVDPSL-NNNSRNEMLRCIHIGLLCVQENLADRPTMTTIMLMLN 1077
Query: 772 SEITHLPPPGRVAFV--HKQSSKSTTESSQKSHQSNSNNNVTLSEVQ 816
S LP P + AF + S S T+S S + + +T+ Q
Sbjct: 1078SYSLSLPIPSKPAFYVSSRTGSISATQSWGYSSGESRSRELTIKSAQ 1218
>TC221391 similar to UP|O81906 (O81906) Serine/threonine kinase-like protein
, partial (25%)
Length = 642
Score = 313 bits (801), Expect = 3e-85
Identities = 153/214 (71%), Positives = 183/214 (85%)
Frame = +2
Query: 520 GQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSL 579
G+E+AVKRLS+ S QG+EEF NE+V+I+KLQHRNLVRLLGCC++ E+ILVYE++PNKSL
Sbjct: 2 GEEVAVKRLSRKSSQGLEEFKNEMVLIAKLQHRNLVRLLGCCIQGEEKILVYEYLPNKSL 181
Query: 580 DAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKI 639
D FLFDP+++ LDW KR IIEGIARG++YLHRDSRLRIIHRDLKASNILLD M PKI
Sbjct: 182 DCFLFDPVKQTQLDWAKRFEIIEGIARGLLYLHRDSRLRIIHRDLKASNILLDESMNPKI 361
Query: 640 SDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRR 699
SDFGLARI G +EANT RVVGTYGYM PEYAMEGLFS KSDVYSFGVLLLEI+SGR+
Sbjct: 362 SDFGLARIFG-GNQNEANTNRVVGTYGYMSPEYAMEGLFSIKSDVYSFGVLLLEIMSGRK 538
Query: 700 NSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEV 733
N+SF +D+ SL+G+AW LW E+ ++ L+DP +
Sbjct: 539 NTSFRDTDDS-SLIGYAWHLWSEQRVMELVDPSL 637
>TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial (23%)
Length = 1167
Score = 286 bits (731), Expect = 3e-77
Identities = 142/243 (58%), Positives = 184/243 (75%), Gaps = 1/243 (0%)
Frame = +2
Query: 576 NKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDM 635
N +L+ + DP + K LDWRKR IIEGI RG++YLHRDSRL+IIHRDLKASN+LL +
Sbjct: 299 NATLNVYFLDPSKSKLLDWRKRCGIIEGIGRGLLYLHRDSRLKIIHRDLKASNVLLYEAL 478
Query: 636 IPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIV 695
PKISDFG+ARI G +D+ANT RVVGTYGYM PEYAM+GLFSEKSDV+SFGVL++EIV
Sbjct: 479 NPKISDFGMARIFG-GTEDQANTNRVVGTYGYMSPEYAMQGLFSEKSDVFSFGVLVIEIV 655
Query: 696 SGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQE 755
SGRRNS F ++ LSL+GFAW W E NI+S+IDPE++D +LRCIHIGLLCVQE
Sbjct: 656 SGRRNSRFYDDDNALSLLGFAWIQWREGNILSVIDPEIYDVTHHKDILRCIHIGLLCVQE 835
Query: 756 LPRDRPNISTVVLMLVSEITHLPPPGRVAFVHKQSSKS-TTESSQKSHQSNSNNNVTLSE 814
DRP ++ V+ ML SE+ LPPP + AFV Q+ + + SS++ + S N +++++
Sbjct: 836 RAVDRPTMAAVISMLNSEVAFLPPPDQPAFVQSQNMLNLVSVSSEERQKLCSINGISITD 1015
Query: 815 VQG 817
++G
Sbjct: 1016IRG 1024
Score = 43.1 bits (100), Expect = 5e-04
Identities = 17/28 (60%), Positives = 22/28 (77%)
Frame = +3
Query: 557 LLGCCVERGEQILVYEFMPNKSLDAFLF 584
L GCC E E++L+YE+M NKSLD F+F
Sbjct: 3 LFGCCAEGDEKMLIYEYMLNKSLDVFIF 86
>TC207570 weakly similar to UP|O65470 (O65470) Serine/threonine kinase-like
protein , partial (30%)
Length = 1285
Score = 99.0 bits (245), Expect(4) = 6e-71
Identities = 54/84 (64%), Positives = 63/84 (74%), Gaps = 1/84 (1%)
Frame = +1
Query: 582 FLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISD 641
F D +K LDW KR NII GIA+G++YLH+ SRL++IHRDLKASNILLD +M KISD
Sbjct: 295 FKTDSARKDLLDWEKRLNIIGGIAQGLLYLHKYSRLKVIHRDLKASNILLDHEMNAKISD 474
Query: 642 FGLARIVKFG-EDDEANTKRVVGT 664
FG+ARI FG E NT RVVGT
Sbjct: 475 FGMARI--FGVRVSEENTNRVVGT 540
Score = 96.7 bits (239), Expect(4) = 6e-71
Identities = 45/75 (60%), Positives = 61/75 (81%)
Frame = +2
Query: 513 YKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYE 572
++G + D QE+A+KRLSK+SGQG+ EF NE +++KLQH NLV+LLG C++R E+ILVYE
Sbjct: 2 HEGNLSDQQEVAIKRLSKSSGQGLIEFTNEAKLMAKLQHTNLVKLLGFCIQRDERILVYE 181
Query: 573 FMPNKSLDAFLFDPL 587
+M NKSLD +LF L
Sbjct: 182 YMSNKSLDFYLFGRL 226
Score = 68.9 bits (167), Expect(4) = 6e-71
Identities = 40/113 (35%), Positives = 65/113 (57%), Gaps = 3/113 (2%)
Frame = +1
Query: 707 EDTLSLVGF--AWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNIS 764
ED L ++ F AW+LW + LID + C ++ + RCIHIGLLCVQ+ DRP +
Sbjct: 832 EDLLWMLEFL*AWQLWNAGRALELIDSTLNGLCSQNEVFRCIHIGLLCVQDQATDRPTMV 1011
Query: 765 TVVLMLVSEITHLPPPGRVA-FVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQ 816
+V L ++ LP P + A F+++ +S +Q+ + +S N+VT+S +
Sbjct: 1012DIVSFLSNDTIQLPQPMQPAYFINEVVEESELPYNQQ--EFHSENDVTISSTR 1164
Score = 64.7 bits (156), Expect(4) = 6e-71
Identities = 29/50 (58%), Positives = 41/50 (82%)
Frame = +2
Query: 666 GYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGF 715
GYM PEYAM+G+ S K+DV+SFGVLLLEI+S ++N+S H + L+L+G+
Sbjct: 629 GYMAPEYAMKGVVSIKTDVFSFGVLLLEILSSKKNNSRYHSDHPLNLIGY 778
>TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (32%)
Length = 738
Score = 260 bits (664), Expect = 2e-69
Identities = 129/219 (58%), Positives = 166/219 (74%)
Frame = +3
Query: 481 DELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFM 540
D+L ++F ++ ATN F N LG+GGFG VYKG + DGQEIA+KRLS S QG EF
Sbjct: 84 DQLLQFEFATIKFATNNFSDANKLGQGGFGIVYKGTLSDGQEIAIKRLSINSNQGETEFK 263
Query: 541 NEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNI 600
NE+++ +LQHRNLVRLLG C R E++L+YEF+PNKSLD F+FDP ++ NL+ R I
Sbjct: 264 NEILLTGRLQHRNLVRLLGFCFARRERLLIYEFVPNKSLDYFIFDPNKRVNLN*EIRYKI 443
Query: 601 IEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKR 660
I GIARG++YLH DSRL ++HRDLK SNILLD ++ PKISDFG+AR+ + + EA+T
Sbjct: 444 IRGIARGLLYLHEDSRLNVVHRDLKTSNILLDGELNPKISDFGMARLFEINQ-TEASTTT 620
Query: 661 VVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRR 699
+VGT+GYM PEY G FS KSDV+SFGV++LEIV G+R
Sbjct: 621 IVGTFGYMAPEYIKHGQFSIKSDVFSFGVMILEIVCGQR 737
>TC208424 weakly similar to UP|O65466 (O65466) Serine/threonine kinase-like
protein , partial (34%)
Length = 913
Score = 256 bits (655), Expect = 2e-68
Identities = 130/248 (52%), Positives = 171/248 (68%)
Frame = +2
Query: 566 EQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLK 625
E+IL+YE++ NKSLD FLFDP+++K LDW +R II GIARGI YLH DS+LRIIHRDLK
Sbjct: 2 EKILIYEYITNKSLDHFLFDPVKQKELDWSRRYKIIVGIARGIQYLHEDSQLRIIHRDLK 181
Query: 626 ASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVY 685
ASN+LLD +M PKISDFG+A+I + + + NT R+VGTYGYM PEYAM G FS KSDV+
Sbjct: 182 ASNVLLDENMNPKISDFGMAKIFQ-ADQTQVNTGRIVGTYGYMSPEYAMRGQFSVKSDVF 358
Query: 686 SFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRC 745
SFGVL+LEIVSG++N+ F L+ AWK W + + L+DP + + + + RC
Sbjct: 359 SFGVLVLEIVSGKKNTDFYQSNHADDLLSHAWKNWTLQTPLELLDPTLRGSYSRNEVNRC 538
Query: 746 IHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSN 805
IHIGLLCVQE P DRP+++T+ LML S + P + A + + + S QS
Sbjct: 539 IHIGLLCVQENPSDRPSMATIALMLNSYSVTMSMPQQPASLLRGRGPNRLNQGMDSDQST 718
Query: 806 SNNNVTLS 813
+N + T S
Sbjct: 719 TNQSTTCS 742
>TC230095 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase
homolog RK20-1, partial (30%)
Length = 819
Score = 254 bits (650), Expect = 8e-68
Identities = 133/272 (48%), Positives = 185/272 (67%), Gaps = 6/272 (2%)
Frame = +3
Query: 544 VVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEG 603
++I+KLQH+NLVRL+G C E E+IL+YE++PNKSLD FLFD + + L W +R I++G
Sbjct: 6 LLIAKLQHKNLVRLVGFCQEDREKILIYEYVPNKSLDHFLFDSQKHRQLTWSERFKIVKG 185
Query: 604 IARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVG 663
IARGI+YLH DSRL+IIHRD+K SN+LLD + PKISDFG+AR+V + + T RVVG
Sbjct: 186 IARGILYLHEDSRLKIIHRDIKPSNVLLDYGINPKISDFGMARMVA-TDQIQGCTNRVVG 362
Query: 664 TYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEE 723
TYGYM PEYAM G FSEKSDV+SFGV++L+I+SG++NS L+ +AW W +E
Sbjct: 363 TYGYMSPEYAMHGQFSEKSDVFSFGVMVLDIISGKKNSCSFESCRVDDLLSYAWNNWRDE 542
Query: 724 NIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRV 783
+ L+D + ++ + + +C+ IGLLCVQE P DRP + T+V L + +P P
Sbjct: 543 SPYQLLDSTLLESYVPNEVEKCMQIGLLCVQENPDDRPTMGTIVSYLSNPSFEMPFPLEP 722
Query: 784 A-FVHKQSSKSTTESSQKS-----HQSNSNNN 809
A F+H + + + E S H S+S+ N
Sbjct: 723 AFFMHGRMRRHSAEHESSSGYYTNHPSSSSVN 818
>TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, partial
(21%)
Length = 765
Score = 251 bits (642), Expect = 7e-67
Identities = 129/226 (57%), Positives = 167/226 (73%), Gaps = 1/226 (0%)
Frame = +3
Query: 592 LDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFG 651
LDW KR NII GIA+G++YLH+DSRLRIIHRDLKASN+LLDS++ PKISDFG+ARI FG
Sbjct: 3 LDWSKRFNIICGIAKGLLYLHQDSRLRIIHRDLKASNVLLDSELNPKISDFGMARI--FG 176
Query: 652 EDD-EANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTL 710
D E NTKR+VGTYGYM PEYA +GLFS KSDV+SFGVLLLEI+SG+R+ + + +
Sbjct: 177 VDQQEGNTKRIVGTYGYMAPEYATDGLFSVKSDVFSFGVLLLEIISGKRSRGYYNQNHSQ 356
Query: 711 SLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 770
+L+G AWKLW E + LID + D+ S ML CIH+ LLCVQ+ P DRP +S+V+LML
Sbjct: 357 NLIGHAWKLWKEGRPLELIDKSIEDSSSLSQMLHCIHVSLLCVQQNPEDRPGMSSVLLML 536
Query: 771 VSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQ 816
VSE+ LP P + F K S ++ + +S++ S + +TL E +
Sbjct: 537 VSEL-ELPEPKQPGFFGKYSGEADSSTSKQQLSSTNEITITLLEAR 671
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 249 bits (637), Expect = 3e-66
Identities = 132/274 (48%), Positives = 183/274 (66%), Gaps = 1/274 (0%)
Frame = +1
Query: 502 NMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCC 561
N +G+GGFGPVYKG DG IAVK+LS S QG EF+NE+ +IS LQH +LV+L GCC
Sbjct: 7 NKIGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQGNREFLNEIGMISALQHPHLVKLYGCC 186
Query: 562 VERGEQILVYEFMPNKSLDAFLFDPLQKK-NLDWRKRSNIIEGIARGIMYLHRDSRLRII 620
VE + +LVYE+M N SL LF + + LDW R I GIARG+ YLH +SRL+I+
Sbjct: 187 VEGDQLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLKIV 366
Query: 621 HRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSE 680
HRD+KA+N+LLD D+ PKISDFGLA++ + ED+ + R+ GT+GYM PEYAM G ++
Sbjct: 367 HRDIKATNVLLDQDLNPKISDFGLAKLDE--EDNTHISTRIAGTFGYMAPEYAMHGYLTD 540
Query: 681 KSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFES 740
K+DVYSFG++ LEI++GR N+ E++ S++ +A L + +I+ L+D + +
Sbjct: 541 KADVYSFGIVALEIINGRSNTIHRQKEESFSVLEWAHLLREKGDIMDLVDRRLGLEFNKE 720
Query: 741 SMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEI 774
L I + LLC RP +S+VV ML +I
Sbjct: 721 EALVMIKVALLCTNVTAALRPTMSSVVSMLEGKI 822
>TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-like kinase
RLK14, partial (20%)
Length = 692
Score = 235 bits (599), Expect = 7e-62
Identities = 126/217 (58%), Positives = 158/217 (72%)
Frame = +3
Query: 597 RSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEA 656
R II GIARG++YLH+DSRLRIIHRDLKASN+LLD++M PKISDFGLAR+ G+ E
Sbjct: 3 RFGIINGIARGLLYLHQDSRLRIIHRDLKASNVLLDNEMNPKISDFGLARMCG-GDQIEG 179
Query: 657 NTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFA 716
T RVVGTYGYM PEYA +G+FS KSDV+SFGVLLLEIVSG++NS + D +L+G A
Sbjct: 180 ETSRVVGTYGYMAPEYAFDGIFSIKSDVFSFGVLLLEIVSGKKNSRLFYPNDYNNLIGHA 359
Query: 717 WKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITH 776
W LW E N + ID + D+C LRCIHIGLLCVQ P DRPN+++VV++L +E
Sbjct: 360 WMLWKEGNPMQFIDSSLEDSCILYEALRCIHIGLLCVQHHPNDRPNMASVVVLLSNE-NA 536
Query: 777 LPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLS 813
LP P +++ K S + ESS K+ S S N+VT+S
Sbjct: 537 LPLPKDPSYLSKDIS-TERESSSKNFTSVSINDVTMS 644
>TC221114 similar to UP|Q8RZ15 (Q8RZ15) Receptor kinase-like protein, partial
(29%)
Length = 946
Score = 234 bits (596), Expect = 1e-61
Identities = 131/270 (48%), Positives = 169/270 (62%), Gaps = 26/270 (9%)
Frame = +3
Query: 422 VRVPAELVAVKKEKGHNKSFLIIVIAGVIGALILVICAYLLWRKCSAR------------ 469
V +P LV K ++LI VIAGV LI ++WRKC
Sbjct: 141 VYIPPGLVHAHHTKSRWWAWLI-VIAGVFVVLIFGYLCCIIWRKCKIEADRKKKQKELLL 317
Query: 470 ------------HKAREHQQM-KLD-ELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKG 515
HK + H++ K++ E+ ++ F + AT F N LG+GGFGPVYKG
Sbjct: 318 EIGVSSVACIVYHKTKRHRKRSKVNYEMQIFSFPIIAAATGNFSVANKLGQGGFGPVYKG 497
Query: 516 VMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMP 575
V+ DGQEIA+KRLS SGQG+ EF NE +++KLQH NLVRL G C++ E IL+YE++P
Sbjct: 498 VLPDGQEIAIKRLSSRSGQGLVEFKNEAELVAKLQHTNLVRLSGLCIQNEENILIYEYLP 677
Query: 576 NKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDM 635
NKSLD LFD +++ + W KR NIIEGIA G++YLH SRL++IHRDLKA NILLD +M
Sbjct: 678 NKSLDFHLFDSKRREKIVWEKRFNIIEGIAHGLIYLHHFSRLKVIHRDLKAGNILLDYEM 857
Query: 636 IPKISDFGLARIVKFGEDDEANTKRVVGTY 665
PKISDFG+A ++ E E TKRVVGTY
Sbjct: 858 NPKISDFGMA-VILDSEVVEVKTKRVVGTY 944
>TC210499 weakly similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine kinase-like
protein , partial (27%)
Length = 717
Score = 231 bits (590), Expect = 7e-61
Identities = 118/209 (56%), Positives = 151/209 (71%), Gaps = 1/209 (0%)
Frame = +3
Query: 592 LDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFG 651
LDW+KR NIIEGI++GI+YLH+ SRL+IIHRDLKASNILLD +M PKISDFGLAR+
Sbjct: 15 LDWKKRFNIIEGISQGILYLHKYSRLKIIHRDLKASNILLDENMNPKISDFGLARMF-MQ 191
Query: 652 EDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLS 711
++ T R+VGTYGYM PEYAMEG FS KSDVYSFGVLLLEIVSGR+N+SF + L+
Sbjct: 192 QESTGTTSRIVGTYGYMSPEYAMEGTFSTKSDVYSFGVLLLEIVSGRKNTSFYDVDHLLN 371
Query: 712 LVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLV 771
L+G AW+LW + + L+DP + D+ + RCIH+GLLCV+ DRP +S V+ ML
Sbjct: 372 LIGHAWELWNQGESLQLLDPSLNDSFDPDEVKRCIHVGLLCVEHYANDRPTMSNVISMLT 551
Query: 772 SEITHLPPPGRVAF-VHKQSSKSTTESSQ 799
+E + P R AF V +++ T S +
Sbjct: 552 NESAPVTLPRRPAFYVERKNFDGKTSSKE 638
>TC210344 similar to UP|Q9AVE0 (Q9AVE0) SRKb, partial (21%)
Length = 870
Score = 231 bits (589), Expect = 1e-60
Identities = 126/220 (57%), Positives = 153/220 (69%)
Frame = +2
Query: 596 KRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDE 655
KR +II GIARGI+YLH DSRL+IIHRDLKASN+LLD DM PKISDFG+ARI G + E
Sbjct: 5 KRLDIINGIARGILYLHEDSRLKIIHRDLKASNVLLDYDMNPKISDFGMARIFA-GSEGE 181
Query: 656 ANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGF 715
ANT +VGTYGYM PEYAMEGL+S KSDV+ FGVLLLEI++G+RN+ F H T SL+ +
Sbjct: 182 ANTATIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGKRNAGFYHSNKTPSLLSY 361
Query: 716 AWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEIT 775
AW LW E + LIDP D+C LR +HIGLLCVQE DRP +S+VVLML +E
Sbjct: 362 AWHLWNEGKGLELIDPMSVDSCPGDEFLRYMHIGLLCVQEDAYDRPTMSSVVLMLKNESA 541
Query: 776 HLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEV 815
L P R F S +++ Q S N +TLS++
Sbjct: 542 TLGQPERPPF-----SLGRFNANEPDCQDCSLNFLTLSDI 646
>TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partial (25%)
Length = 1190
Score = 230 bits (586), Expect = 2e-60
Identities = 125/253 (49%), Positives = 170/253 (66%)
Frame = +3
Query: 522 EIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDA 581
EIAVK+LS S QG +F+ E+ IS +QHRNLV+L GCC+E +++LVYE++ NKSLD
Sbjct: 3 EIAVKQLSVGSHQGKSQFITEIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQ 182
Query: 582 FLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISD 641
LF L+W R +I G+ARG+ YLH +SRLRI+HRD+KASNILLD ++IPKISD
Sbjct: 183 ALFGKCL--TLNWSTRYDICLGVARGLTYLHEESRLRIVHRDVKASNILLDYELIPKISD 356
Query: 642 FGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNS 701
FGLA++ + + + V GT GY+ PEYAM G +EK+DV+SFGV+ LE+VSGR NS
Sbjct: 357 FGLAKL--YDDKKTHISTGVAGTIGYLAPEYAMRGHLTEKADVFSFGVVALELVSGRPNS 530
Query: 702 SFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRP 761
S + + L+ +AW+L + II L+D + E + R + I LLC Q P RP
Sbjct: 531 DSSLEGEKVYLLEWAWQLHEKNCIIDLVDDRL-SEFNEEEVKRVVGIALLCTQTSPTLRP 707
Query: 762 NISTVVLMLVSEI 774
++S VV ML +I
Sbjct: 708 SMSRVVAMLSGDI 746
>TC213718 similar to UP|Q70I29 (Q70I29) S-receptor kinase-like protein 2,
partial (20%)
Length = 525
Score = 227 bits (579), Expect = 1e-59
Identities = 112/174 (64%), Positives = 138/174 (78%)
Frame = +1
Query: 596 KRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDE 655
KR II+GIARG++YLH+DSRLRIIHRDLK SNILLD+DM PKISDFGLAR G+ E
Sbjct: 7 KRLQIIDGIARGLLYLHQDSRLRIIHRDLKVSNILLDNDMNPKISDFGLARTFG-GDQAE 183
Query: 656 ANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGF 715
ANT RV+GTYGYMPPEYA+ G FS KSDV+SFGV++LEI+SGR+N +F E L+L+
Sbjct: 184 ANTNRVMGTYGYMPPEYALHGRFSIKSDVFSFGVIVLEIISGRKNRNFQDSEHHLNLLSH 363
Query: 716 AWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLM 769
AW+LW+EE + LID + D +LRCIH+GLLCVQ+ P +RPN+S+VVLM
Sbjct: 364 AWRLWIEEKPLELIDDLLDDPVSPHEILRCIHVGLLCVQQTPENRPNMSSVVLM 525
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,415,557
Number of Sequences: 63676
Number of extensions: 647218
Number of successful extensions: 5453
Number of sequences better than 10.0: 1009
Number of HSP's better than 10.0 without gapping: 4566
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4641
length of query: 817
length of database: 12,639,632
effective HSP length: 105
effective length of query: 712
effective length of database: 5,953,652
effective search space: 4239000224
effective search space used: 4239000224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144721.7


