

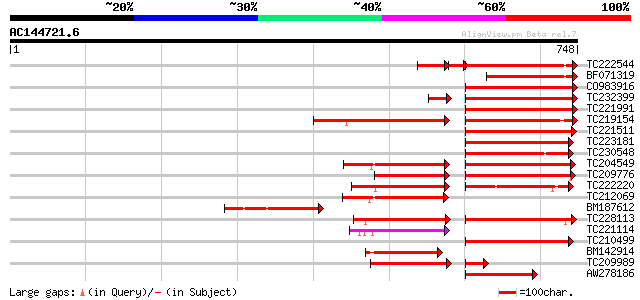
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144721.6 + phase: 0 /pseudo
(748 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precu... 211 3e-56
BF071319 178 9e-45
CO983916 171 1e-42
TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial... 167 2e-41
TC221991 weakly similar to UP|O23743 (O23743) SFR1 protein precu... 159 6e-39
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 151 1e-36
TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, ... 147 1e-35
TC223181 similar to UP|Q40096 (Q40096) Receptor protein kinase, ... 141 9e-34
TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-l... 137 2e-32
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 137 2e-32
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 134 1e-31
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 133 3e-31
TC212069 weakly similar to UP|Q9M0X5 (Q9M0X5) Receptor protein k... 132 4e-31
BM187612 132 4e-31
TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 132 4e-31
TC221114 similar to UP|Q8RZ15 (Q8RZ15) Receptor kinase-like prot... 132 4e-31
TC210499 weakly similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine k... 132 6e-31
BM142914 similar to GP|9294449|dbj| receptor kinase 1 {Arabidops... 131 1e-30
TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 130 2e-30
AW278186 weakly similar to GP|4008010|gb| receptor-like protein ... 124 2e-28
>TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precursor ,
partial (27%)
Length = 861
Score = 211 bits (537), Expect(2) = 3e-56
Identities = 107/147 (72%), Positives = 122/147 (82%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAMEG+FSEKSDVY VLLLEIVSGRRN+SFY+NE SLSL +AWKLW E NI S+I
Sbjct: 387 PEYAMEGIFSEKSDVYXXXVLLLEIVSGRRNTSFYNNEQSLSLXXYAWKLWNEGNIKSII 566
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D E+ D FE S+LRCIHIGLLCVQEL ++RP ISTVV MLISEITHLPPP + FV K+
Sbjct: 567 DLEIQDPMFEKSILRCIHIGLLCVQELTKERPTISTVVXMLISEITHLPPPRQXXFVQKQ 746
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
N +S ESSQKS Q NSNN+VT+SE+QG
Sbjct: 747 NCQSSESSQKS-QFNSNNNVTISEIQG 824
Score = 84.0 bits (206), Expect = 2e-16
Identities = 39/43 (90%), Positives = 41/43 (94%)
Frame = +3
Query: 538 EVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
EV VISKLQHRNLVRLLGCC+ER EQMLVYEFMPNKSLD+FLF
Sbjct: 3 EVVVISKLQHRNLVRLLGCCIERDEQMLVYEFMPNKSLDSFLF 131
Score = 26.9 bits (58), Expect(2) = 3e-56
Identities = 15/26 (57%), Positives = 18/26 (68%)
Frame = +2
Query: 580 FG*NC*RRRR**NKHK*GCWNLPEYA 605
F *NC*+ **+ H+ GCWNL YA
Sbjct: 308 FS*NC*KW****S*HEKGCWNLWLYA 385
>BF071319
Length = 420
Score = 178 bits (451), Expect = 9e-45
Identities = 88/119 (73%), Positives = 103/119 (85%)
Frame = +1
Query: 630 RRNSSFYHNEDSLSLVGFAWKLWLEENIISLIDREVWDASFESSMLRCIHIGLLCVQELP 689
RRN+SFY+NE SLSLVG+AWKLW E+NI+S+ID E+ D FE S+LRCIHIGLLCVQEL
Sbjct: 1 RRNTSFYNNEQSLSLVGYAWKLWNEDNIMSIIDPEIHDPMFEKSILRCIHIGLLCVQELT 180
Query: 690 RDRPNISTVVLMLISEITHLPPPGKVAFVHKKNSKSGESSQKSQQSNSNNSVTLSEVQG 748
++RP ISTVVLMLISEITHLPPP +VAFV K+N +S ESSQKS Q NSNN VT+SE+QG
Sbjct: 181 KERPTISTVVLMLISEITHLPPPRQVAFVQKQNCQSSESSQKS-QFNSNNDVTISEIQG 354
>CO983916
Length = 835
Score = 171 bits (433), Expect = 1e-42
Identities = 83/147 (56%), Positives = 111/147 (75%)
Frame = -1
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM+GLFSEKSDV+SFGVL+LEIVSGRRNSSFY NE+ LSL+GFAW W E NI+SL+
Sbjct: 751 PEYAMQGLFSEKSDVFSFGVLVLEIVSGRRNSSFYDNENFLSLLGFAWIQWKEGNILSLV 572
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D +D S+ +LRCIHIG LCVQEL +RP ++TV+ ML S+ LPPP + AF+ ++
Sbjct: 571 DPGTYDPSYHKEILRCIHIGFLCVQELAVERPTMATVISMLNSDDVFLPPPSQPAFILRQ 392
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
N + SS++ S N+V+++++ G
Sbjct: 391 NMLNSVSSEEIHNFVSINTVSITDIHG 311
>TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial (23%)
Length = 1167
Score = 167 bits (423), Expect = 2e-41
Identities = 81/149 (54%), Positives = 113/149 (75%), Gaps = 2/149 (1%)
Frame = +2
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM+GLFSEKSDV+SFGVL++EIVSGRRNS FY ++++LSL+GFAW W E NI+S+I
Sbjct: 578 PEYAMQGLFSEKSDVFSFGVLVIEIVSGRRNSRFYDDDNALSLLGFAWIQWREGNILSVI 757
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D E++D + +LRCIHIGLLCVQE DRP ++ V+ ML SE+ LPPP + AFV +
Sbjct: 758 DPEIYDVTHHKDILRCIHIGLLCVQERAVDRPTMAAVISMLNSEVAFLPPPDQPAFVQSQ 937
Query: 722 NSKS--GESSQKSQQSNSNNSVTLSEVQG 748
N + SS++ Q+ S N +++++++G
Sbjct: 938 NMLNLVSVSSEERQKLCSINGISITDIRG 1024
Score = 47.4 bits (111), Expect = 2e-05
Identities = 20/30 (66%), Positives = 24/30 (79%)
Frame = +3
Query: 553 LLGCCVERGEQMLVYEFMPNKSLDAFLFG* 582
L GCC E E+ML+YE+M NKSLD F+FG*
Sbjct: 3 LFGCCAEGDEKMLIYEYMLNKSLDVFIFG* 92
>TC221991 weakly similar to UP|O23743 (O23743) SFR1 protein precursor ,
partial (13%)
Length = 776
Score = 159 bits (401), Expect = 6e-39
Identities = 79/147 (53%), Positives = 112/147 (75%)
Frame = +2
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA GL SEK DV+SFGVLLLEI+SGR+ SS+Y ++ S+SL+GFAWKLW E++I S+I
Sbjct: 134 PEYAFRGLVSEKLDVFSFGVLLLEIISGRKISSYYDHDQSMSLLGFAWKLWNEKDIQSVI 313
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D E+ + + + + RCIHIGLLC+Q L +RP ++TVV ML SEI +LP P AFV ++
Sbjct: 314 DPEISNPNHVNDIERCIHIGLLCLQNLATERPIMATVVSMLNSEIVNLPRPSHPAFVDRQ 493
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
S ESS+++ ++ S N+VT++++QG
Sbjct: 494 IVSSAESSRQNHRTQSINNVTVTDMQG 574
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 151 bits (381), Expect = 1e-36
Identities = 83/187 (44%), Positives = 126/187 (66%), Gaps = 8/187 (4%)
Frame = +1
Query: 402 CMYWSSELIDLQKFPT--SGVDLFIRVP-AELEKEKGNKSFLII---AIAGGLGAFILVI 455
C+ W +L+D++ + SG L IR+P +ELE K KS II ++A LG + +
Sbjct: 49 CVMWFGDLLDIKLYSVAESGRRLHIRLPPSELESIKSKKSSKIIIGTSVAAPLGVVLAIC 228
Query: 456 CAYL--LWRKWSARHTEQKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGIL 513
Y + K + + ++++ ++PL+D + + AT++F +N +G+GGFGPVYKG L
Sbjct: 229 FIYRRNIADKSKTKKSIDRQLQDVDVPLFDMLTITAATDNFLLNNKIGEGGFGPVYKGKL 408
Query: 514 EDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNK 573
GQE+AVKRLS SGQGI EF+ EV +I+KLQHRNLV+LLGCC++ E++LVYE++ N
Sbjct: 409 VGGQEIAVKRLSSLSGQGITEFITEVKLIAKLQHRNLVKLLGCCIKGQEKLLVYEYVVNG 588
Query: 574 SLDAFLF 580
SL++F+F
Sbjct: 589 SLNSFIF 609
Score = 127 bits (319), Expect = 2e-29
Identities = 66/147 (44%), Positives = 94/147 (63%)
Frame = +1
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA +G FS KSDV+SFG+LLLEIV G +N S H +L+LVG+AW LW E+N + LI
Sbjct: 862 PEYAFDGNFSIKSDVFSFGILLLEIVCGIKNKSLCHENQTLNLVGYAWALWKEQNALQLI 1041
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D + D+ +LRCIH+ LLCVQ+ P DRP +++V+ ML SE+ + P + F ++
Sbjct: 1042 DSGIKDSCVIPEVLRCIHVSLLCVQQYPEDRPTMTSVIQMLGSEM-DMVEPKEPGFFPRR 1218
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
K G ++ SN+ +T+S G
Sbjct: 1219 ILKEG----NLKEMTSNDELTISLFSG 1287
>TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, partial
(21%)
Length = 765
Score = 147 bits (372), Expect = 1e-35
Identities = 74/147 (50%), Positives = 104/147 (70%), Gaps = 1/147 (0%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA +GLFS KSDV+SFGVLLLEI+SG+R+ +Y+ S +L+G AWKLW E + LI
Sbjct: 234 PEYATDGLFSVKSDVFSFGVLLLEIISGKRSRGYYNQNHSQNLIGHAWKLWKEGRPLELI 413
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D+ + D+S S ML CIH+ LLCVQ+ P DRP +S+V+LML+SE+ LP P + F K
Sbjct: 414 DKSIEDSSSLSQMLHCIHVSLLCVQQNPEDRPGMSSVLLMLVSEL-ELPEPKQPGFFGKY 590
Query: 722 NSKSGESSQKSQQSNSNN-SVTLSEVQ 747
+ ++ S+ K Q S++N ++TL E +
Sbjct: 591 SGEADSSTSKQQLSSTNEITITLLEAR 671
>TC223181 similar to UP|Q40096 (Q40096) Receptor protein kinase, partial
(17%)
Length = 565
Score = 141 bits (356), Expect = 9e-34
Identities = 71/144 (49%), Positives = 99/144 (68%), Gaps = 1/144 (0%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM+G+FS KSDV+SFGVL+LEI+SG++N FY L+L+G AWKLW EEN + LI
Sbjct: 9 PEYAMDGIFSVKSDVFSFGVLVLEIISGKKNRGFYSANKELNLLGHAWKLWKEENALELI 188
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D + ++ ES +LRCI +GLLCVQE DRP +++VVLML S+ + P F +
Sbjct: 189 DPSIDNSYSESEVLRCIQVGLLCVQERAEDRPTMASVVLMLSSDTASMSQPKNPGFCLGR 368
Query: 722 N-SKSGESSQKSQQSNSNNSVTLS 744
N ++ SS K ++S + N VT++
Sbjct: 369 NPMETDSSSSKQEESCTVNQVTVT 440
>TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-like kinase
RLK14, partial (20%)
Length = 692
Score = 137 bits (344), Expect = 2e-32
Identities = 73/143 (51%), Positives = 96/143 (67%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA +G+FS KSDV+SFGVLLLEIVSG++NS ++ D +L+G AW LW E N + I
Sbjct: 219 PEYAFDGIFSIKSDVFSFGVLLLEIVSGKKNSRLFYPNDYNNLIGHAWMLWKEGNPMQFI 398
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D + D+ LRCIHIGLLCVQ P DRPN+++VV++L +E LP P +++ K
Sbjct: 399 DSSLEDSCILYEALRCIHIGLLCVQHHPNDRPNMASVVVLLSNE-NALPLPKDPSYLSKD 575
Query: 722 NSKSGESSQKSQQSNSNNSVTLS 744
S ESS K+ S S N VT+S
Sbjct: 576 ISTERESSSKNFTSVSINDVTMS 644
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 137 bits (344), Expect = 2e-32
Identities = 77/150 (51%), Positives = 97/150 (64%), Gaps = 10/150 (6%)
Frame = +3
Query: 441 IIAIAGGLGAFILVICAYLLWRKWSARHTEQKEMK----------LDELPLYDFVKLENA 490
I+AI + +L+ + + AR +Q +K +D L +DF +E A
Sbjct: 642 IVAIVVPITVAVLIFIVGICFLSRRARKKQQGSVKEGKTAYDIPTVDSLQ-FDFSTIEAA 818
Query: 491 TNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNL 550
TN F N LG+GGFG VYKG L GQ VAVKRLSKSSGQG EEF NEV V++KLQHRNL
Sbjct: 819 TNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRLSKSSGQGGEEFKNEVVVVAKLQHRNL 998
Query: 551 VRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
VRLLG C++ E++LVYE++PNKSLD LF
Sbjct: 999 VRLLGFCLQGEEKILVYEYVPNKSLDYILF 1088
Score = 127 bits (319), Expect = 2e-29
Identities = 69/148 (46%), Positives = 97/148 (64%), Gaps = 3/148 (2%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM G FS KSDVYSFGVLL+EI+SG++NSSFY + + L+ +AW+LW + + L+
Sbjct: 1341 PEYAMHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTDGAEDLLSYAWQLWKDGTPLELM 1520
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVA-FVHK 720
D + ++ ++ ++R IHIGLLCVQE P DRP ++T+VLML S LP P + A FVH
Sbjct: 1521 DPILRESYNQNEVIRSIHIGLLCVQEDPADRPTMATIVLMLDSNTVTLPTPTQPAFFVHS 1700
Query: 721 KNSKSGESSQKSQQS--NSNNSVTLSEV 746
+ QS S N +++SE+
Sbjct: 1701 GTDPNMPKELPFDQSIPMSVNDMSISEM 1784
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 134 bits (337), Expect = 1e-31
Identities = 71/152 (46%), Positives = 101/152 (65%), Gaps = 7/152 (4%)
Frame = +2
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM G +S KSDVYSFGVL+LEI+SG+RNSSFY + + L+ +AWKLW +E + L+
Sbjct: 593 PEYAMHGEYSAKSDVYSFGVLILEIISGKRNSSFYETDVAEDLLSYAWKLWKDEAPLELM 772
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAF-VHK 720
D+ + ++ + ++RCIHIGLLCVQE P DRP +++VVLML S L P + AF ++
Sbjct: 773 DQSLRESYTRNEVIRCIHIGLLCVQEDPIDRPTMASVVLMLDSYSVTLQVPNQPAFYINS 952
Query: 721 KNSKSGESSQKSQQSNSN------NSVTLSEV 746
+ + K QS +N N +++SEV
Sbjct: 953 RTEPNMPKGLKIDQSTTNSTSKSVNDMSVSEV 1048
Score = 133 bits (335), Expect = 3e-31
Identities = 66/99 (66%), Positives = 78/99 (78%)
Frame = +2
Query: 482 YDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAV 541
+DF +E AT F +N LG+GGFG VYKG+L GQEVAVKRLSK SGQG EEF NEV +
Sbjct: 44 FDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNEVEI 223
Query: 542 ISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
++KLQHRNLVRLLG C+E E++LVYEF+ NKSLD LF
Sbjct: 224 VAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILF 340
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 133 bits (334), Expect = 3e-31
Identities = 69/134 (51%), Positives = 91/134 (67%), Gaps = 5/134 (3%)
Frame = +3
Query: 452 ILVICAYLLWRKWSARHTEQKEMKLDELPL-----YDFVKLENATNSFHNSNMLGKGGFG 506
+L +C ++L R +T +E +E + V +E ATN F +G+GGFG
Sbjct: 84 LLCVCYFILKRSRKKYNTLLRENFGEESATLESLQFGLVTIEAATNKFSYEKRIGEGGFG 263
Query: 507 PVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLV 566
VYKG+L DG+E+AVK+LSKSSGQG EF NE+ +I+KLQHRNLV LLG C+E E+ML+
Sbjct: 264 VVYKGVLPDGREIAVKKLSKSSGQGANEFKNEILLIAKLQHRNLVTLLGFCLEEHEKMLI 443
Query: 567 YEFMPNKSLDAFLF 580
YEF+ NKSLD FLF
Sbjct: 444 YEFVSNKSLDYFLF 485
Score = 110 bits (275), Expect = 2e-24
Identities = 67/149 (44%), Positives = 94/149 (62%), Gaps = 7/149 (4%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNS-SFYHNEDSLSLVGFAWKLWLEENIISL 660
PEYAM G FSEKSDV+SFGV++LEI+S +RN+ S + + D L+ + W+ W++E +++
Sbjct: 738 PEYAMHGQFSEKSDVFSFGVIVLEIISAKRNTRSVFSDHD--DLLSYTWEQWMDEAPLNI 911
Query: 661 IDREVWDASF--ESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGK---- 714
D+ + +A F S +++CI IGLLCVQE P DRP I+ V+ L S IT LP P K
Sbjct: 912 FDQSI-EAEFCDHSEVVKCIQIGLLCVQEKPDDRPKITQVISYLNSSITELPLPKKPIRQ 1088
Query: 715 VAFVHKKNSKSGESSQKSQQSNSNNSVTL 743
V K GESS S S + SV++
Sbjct: 1089SGIVQK--IAVGESSSGSTPSINEMSVSI 1169
>TC212069 weakly similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like
protein, partial (19%)
Length = 513
Score = 132 bits (333), Expect = 4e-31
Identities = 74/146 (50%), Positives = 95/146 (64%), Gaps = 6/146 (4%)
Frame = +1
Query: 440 LIIAIAGGLGAFILVICAYLLWRKWSARHTEQKE------MKLDELPLYDFVKLENATNS 493
+++ I L L C +L + +H KE L+ L ++ K+E ATN
Sbjct: 7 IVVPIVVSLVLLSLGCCCFLHRKATKNQHDILKENFGNDSTTLETLR-FELAKIEAATNR 183
Query: 494 FHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRL 553
F NM+GKGGFG VY+GIL DGQE+AVKRL+ SS QG EF NEV VI+KLQHRNLVRL
Sbjct: 184 FAKENMIGKGGFGEVYRGILLDGQEIAVKRLTGSSRQGAVEFKNEVQVIAKLQHRNLVRL 363
Query: 554 LGCCVERGEQMLVYEFMPNKSLDAFL 579
G C+E E++L+YE++PNKSLD FL
Sbjct: 364 QGFCLEDDEKILIYEYVPNKSLDYFL 441
>BM187612
Length = 423
Score = 132 bits (333), Expect = 4e-31
Identities = 66/132 (50%), Positives = 83/132 (62%), Gaps = 2/132 (1%)
Frame = +3
Query: 284 TVSQNECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLK 343
T ++CD YG CG F C+ S P ICSC KGFE +N EW+ +NWT GCVR+ L+
Sbjct: 36 TSQDSDCDVYGICGSFAICNAQSSP-ICSCLKGFEARNKEEWNRQNWTGGCVRRT--QLQ 206
Query: 344 CEMVK--NGSSVVKQDKFLVHPNTKPPDFAERSDVSRDKCRTDCLANCSCLAYAYDPFIR 401
CE VK N S+ K+D FL K P FAE S V D CR+ CL NCSC+AY++D I
Sbjct: 207 CERVKDHNTSTDTKEDGFLKLQMVKVPYFAEGSPVEPDICRSQCLENCSCVAYSHDDGIG 386
Query: 402 CMYWSSELIDLQ 413
CM W+ L+D+Q
Sbjct: 387 CMSWTGNLLDIQ 422
>TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (53%)
Length = 1492
Score = 132 bits (333), Expect = 4e-31
Identities = 67/135 (49%), Positives = 93/135 (68%), Gaps = 7/135 (5%)
Frame = +1
Query: 454 VICAYLLWRKWSAR-------HTEQKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFG 506
+IC + R+ AR H + E+++ + ++F + AT F +SN LG+GGFG
Sbjct: 13 LICLCIYSRRSKARKSSLVKQHEDDDEIEIAQSLQFNFDTIRVATEDFSDSNKLGQGGFG 192
Query: 507 PVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLV 566
VY+G L DGQ +AVKRLS+ S QG EF NEV +++KLQHRNLVRLLG C+E E++L+
Sbjct: 193 AVYRGRLSDGQMIAVKRLSRESSQGDTEFKNEVLLVAKLQHRNLVRLLGFCLEGKERLLI 372
Query: 567 YEFMPNKSLDAFLFG 581
YE++PNKSLD F+FG
Sbjct: 373 YEYVPNKSLDYFIFG 417
Score = 122 bits (307), Expect = 4e-28
Identities = 67/149 (44%), Positives = 91/149 (60%), Gaps = 3/149 (2%)
Frame = +1
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM G FS KSDV+SFGVL+LEI+SG +NS H E+ L+ FAW+ W E + ++
Sbjct: 775 PEYAMHGQFSMKSDVFSFGVLVLEIISGHKNSGIRHGENVEDLLSFAWRNWREGTAVKIV 954
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D + + S + MLRCIHIGLLCVQE DRP ++T++LML S LP P K AF
Sbjct: 955 DPSLNNNS-RNEMLRCIHIGLLCVQENLADRPTMTTIMLMLNSYSLSLPIPSKPAFYVSS 1131
Query: 722 NSKSGESSQK---SQQSNSNNSVTLSEVQ 747
+ S ++Q S + + +T+ Q
Sbjct: 1132RTGSISATQSWGYSSGESRSRELTIKSAQ 1218
>TC221114 similar to UP|Q8RZ15 (Q8RZ15) Receptor kinase-like protein, partial
(29%)
Length = 946
Score = 132 bits (333), Expect = 4e-31
Identities = 71/163 (43%), Positives = 98/163 (59%), Gaps = 31/163 (19%)
Frame = +3
Query: 449 GAFILVICAYL---LWRKWSA---RHTEQKEMKLD------------------------- 477
G F+++I YL +WRK R +QKE+ L+
Sbjct: 216 GVFVVLIFGYLCCIIWRKCKIEADRKKKQKELLLEIGVSSVACIVYHKTKRHRKRSKVNY 395
Query: 478 ELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMN 537
E+ ++ F + AT +F +N LG+GGFGPVYKG+L DGQE+A+KRLS SGQG+ EF N
Sbjct: 396 EMQIFSFPIIAAATGNFSVANKLGQGGFGPVYKGVLPDGQEIAIKRLSSRSGQGLVEFKN 575
Query: 538 EVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
E +++KLQH NLVRL G C++ E +L+YE++PNKSLD LF
Sbjct: 576 EAELVAKLQHTNLVRLSGLCIQNEENILIYEYLPNKSLDFHLF 704
>TC210499 weakly similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine kinase-like
protein , partial (27%)
Length = 717
Score = 132 bits (332), Expect = 6e-31
Identities = 67/143 (46%), Positives = 93/143 (64%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAMEG FS KSDVYSFGVLLLEIVSGR+N+SFY + L+L+G AW+LW + + L+
Sbjct: 246 PEYAMEGTFSTKSDVYSFGVLLLEIVSGRKNTSFYDVDHLLNLIGHAWELWNQGESLQLL 425
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D + D+ + RCIH+GLLCV+ DRP +S V+ ML +E + P + AF ++
Sbjct: 426 DPSLNDSFDPDEVKRCIHVGLLCVEHYANDRPTMSNVISMLTNESAPVTLPRRPAFYVER 605
Query: 722 NSKSGESSQKSQQSNSNNSVTLS 744
+ G++S K +S + T S
Sbjct: 606 KNFDGKTSSKELCVDSTDEFTAS 674
>BM142914 similar to GP|9294449|dbj| receptor kinase 1 {Arabidopsis
thaliana}, partial (11%)
Length = 413
Score = 131 bits (330), Expect = 1e-30
Identities = 64/101 (63%), Positives = 81/101 (79%)
Frame = +1
Query: 470 EQKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSG 529
++KE+KL PL+ FV + ATN+F ++N LG+GGFGPVYKGIL +G EVAVKRLS+ SG
Sbjct: 118 KKKEVKL---PLFSFVSVAAATNNFSDANKLGEGGFGPVYKGILLNGDEVAVKRLSRRSG 288
Query: 530 QGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFM 570
QG EE NE +I+KLQH NLVRLLGCC++R E+ML+YE M
Sbjct: 289 QGWEELRNEALLIAKLQHNNLVRLLGCCIDRDEKMLIYELM 411
>TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (32%)
Length = 738
Score = 130 bits (328), Expect = 2e-30
Identities = 62/107 (57%), Positives = 82/107 (75%)
Frame = +3
Query: 477 DELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFM 536
D+L ++F ++ ATN+F ++N LG+GGFG VYKG L DGQE+A+KRLS +S QG EF
Sbjct: 84 DQLLQFEFATIKFATNNFSDANKLGQGGFGIVYKGTLSDGQEIAIKRLSINSNQGETEFK 263
Query: 537 NEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLFG*N 583
NE+ + +LQHRNLVRLLG C R E++L+YEF+PNKSLD F+F N
Sbjct: 264 NEILLTGRLQHRNLVRLLGFCFARRERLLIYEFVPNKSLDYFIFDPN 404
Score = 43.9 bits (102), Expect = 3e-04
Identities = 20/30 (66%), Positives = 24/30 (79%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRR 631
PEY G FS KSDV+SFGV++LEIV G+R
Sbjct: 648 PEYIKHGQFSIKSDVFSFGVMILEIVCGQR 737
>AW278186 weakly similar to GP|4008010|gb| receptor-like protein kinase
{Arabidopsis thaliana}, partial (16%)
Length = 429
Score = 124 bits (311), Expect = 2e-28
Identities = 56/95 (58%), Positives = 76/95 (79%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAMEGLFSEKSD+YSFGVLLLEI+SG+RN+S +++ SLSL+G+AW LW E+NI L+
Sbjct: 144 PEYAMEGLFSEKSDIYSFGVLLLEIISGKRNTSCRNDDQSLSLIGYAWNLWNEDNISFLV 323
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNIS 696
D E+ + E+ + RCIHI +CVQE+ + RP ++
Sbjct: 324 DPEISASGSENHIFRCIHIAFVCVQEVAKTRPTMT 428
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,529,820
Number of Sequences: 63676
Number of extensions: 584535
Number of successful extensions: 4275
Number of sequences better than 10.0: 797
Number of HSP's better than 10.0 without gapping: 3876
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4037
length of query: 748
length of database: 12,639,632
effective HSP length: 104
effective length of query: 644
effective length of database: 6,017,328
effective search space: 3875159232
effective search space used: 3875159232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144721.6


