

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144720.7 + phase: 2 /pseudo/partial
(1007 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
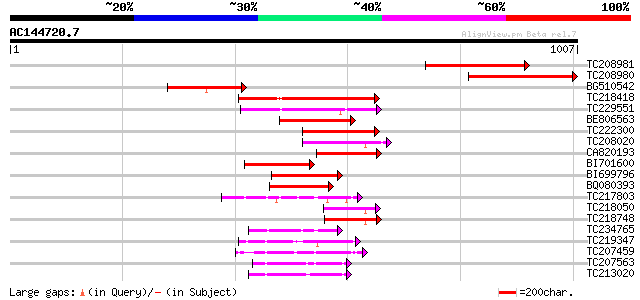
Score E
Sequences producing significant alignments: (bits) Value
TC208981 similar to UP|Q9FF46 (Q9FF46) ABC transporter-like prot... 344 1e-94
TC208980 similar to UP|Q9FF46 (Q9FF46) ABC transporter-like prot... 339 3e-93
BG510542 weakly similar to GP|9759338|dbj ABC transporter-like p... 192 6e-49
TC218418 homologue to UP|Q6X4V5 (Q6X4V5) ABC transporter, partia... 186 3e-47
TC229551 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, p... 166 5e-41
BE806563 similar to GP|11994752|db ABC transporter-like protein ... 123 4e-28
TC222300 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {A... 106 6e-23
TC208020 similar to UP|Q76CU2 (Q76CU2) PDR-type ABC transporter ... 99 9e-21
CA820193 similar to PIR|T04229|T04 ABC-type transport protein F1... 94 3e-19
BI701600 93 5e-19
BI699796 similar to PIR|G85167|G85 ABC transporter like protein ... 90 6e-18
BQ080393 84 3e-16
TC217803 similar to UP|Q9AT00 (Q9AT00) At1g65410/T8F5_19, partia... 75 1e-13
TC218050 weakly similar to UP|Q7PC82 (Q7PC82) PDR14 ABC transpor... 74 4e-13
TC218748 similar to UP|Q76CU1 (Q76CU1) PDR-type ABC transporter ... 72 9e-13
TC234765 similar to UP|Q84M24 (Q84M24) AtABCA1, partial (9%) 72 2e-12
TC219347 similar to UP|Q9LF78 (Q9LF78) Mitochondrial half-ABC tr... 71 3e-12
TC207459 similar to UP|Q94IH6 (Q94IH6) CjMDR1, partial (30%) 70 5e-12
TC207563 similar to UP|Q9ZRG2 (Q9ZRG2) P-glycoprotein, partial (... 68 2e-11
TC213020 weakly similar to UP|Q8GU71 (Q8GU71) MDR-like ABC trans... 67 3e-11
>TC208981 similar to UP|Q9FF46 (Q9FF46) ABC transporter-like protein, partial
(16%)
Length = 555
Score = 344 bits (883), Expect = 1e-94
Identities = 169/184 (91%), Positives = 178/184 (95%)
Frame = +3
Query: 739 RLNFLKSKDLSNRKTPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKS 798
RLNF KSKDLSNR+TPGVFKQYKYFLIRVGKQRLREARIQA+DYLILLLAGACLGS++KS
Sbjct: 3 RLNFFKSKDLSNRETPGVFKQYKYFLIRVGKQRLREARIQAIDYLILLLAGACLGSLSKS 182
Query: 799 SDQTFGASGYTYTVIAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDHFN 858
SDQTFGA+GYT+TVI VSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDT+DHFN
Sbjct: 183 SDQTFGAAGYTHTVIGVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTIDHFN 362
Query: 859 TVIKPVVYLSMFYFLTNPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPGAAQLWSVLLP 918
T+IKPVVYLSMFYF TNPRSTF DNY+VLLCLVYCVTGIAYALSI FEPGAAQLWSVLLP
Sbjct: 363 TLIKPVVYLSMFYFFTNPRSTFADNYVVLLCLVYCVTGIAYALSIFFEPGAAQLWSVLLP 542
Query: 919 VVST 922
VV T
Sbjct: 543 VVLT 554
>TC208980 similar to UP|Q9FF46 (Q9FF46) ABC transporter-like protein, partial
(15%)
Length = 820
Score = 339 bits (870), Expect = 3e-93
Identities = 168/193 (87%), Positives = 175/193 (90%)
Frame = +1
Query: 815 VSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDHFNTVIKPVVYLSMFYFLT 874
V LCK AA S LDKLHY ESDSGMSSLAYFLSKDT+DH NT+IKPVVYLSMFYF T
Sbjct: 4 VXXLCKXAAXXSXXLDKLHYGGESDSGMSSLAYFLSKDTIDHXNTLIKPVVYLSMFYFFT 183
Query: 875 NPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPGAAQLWSVLLPVVSTLIATQQKDSKIL 934
NP STF DNY+VLLCLVYCVTGIAYALSI FEPGAAQLWSVLLPVV TLIATQ KDSK+L
Sbjct: 184 NPISTFADNYVVLLCLVYCVTGIAYALSIFFEPGAAQLWSVLLPVVLTLIATQPKDSKVL 363
Query: 935 KAIANLCYSKWALQALVIANAERYQGVWLITRCGSLLKSGYNLHDWSLCISILILMGVIG 994
K IANLCYSKWALQALV+ANAERYQGVWLITRCGSLLK+GYNLHDWSLCISILILMGVI
Sbjct: 364 KNIANLCYSKWALQALVVANAERYQGVWLITRCGSLLKTGYNLHDWSLCISILILMGVIC 543
Query: 995 RAIAFFCMVTFKK 1007
RAIAFFCMVTF+K
Sbjct: 544 RAIAFFCMVTFRK 582
>BG510542 weakly similar to GP|9759338|dbj ABC transporter-like protein
{Arabidopsis thaliana}, partial (6%)
Length = 439
Score = 192 bits (488), Expect = 6e-49
Identities = 102/146 (69%), Positives = 118/146 (79%), Gaps = 6/146 (4%)
Frame = -1
Query: 281 KVKILNQETSETDVELLPHSQP--SNMVASSSAVPTEKGKTPSGLMHMMHEIENDPHVNY 338
KVKILNQ TSE D+ELL HS P S+MVASSS P EKGK P+GLM ++HEIENDP +N
Sbjct: 439 KVKILNQATSEADIELLSHSGPTTSSMVASSSLAPKEKGKEPNGLMQIIHEIENDPDIND 260
Query: 339 NPNTGKETR----HKSATKEKQPQTNTQIFKYAYAQLEKEKAQQQENKNLTFSGVLKMAT 394
+ +T ETR +A K KQP T++QIFKYAY+QLEKEKA+QQENK LTFSGV+KMAT
Sbjct: 259 HLHTEIETRDTGVRANAPKGKQPHTHSQIFKYAYSQLEKEKAEQQENKKLTFSGVIKMAT 80
Query: 395 NTEKSKRPFIEISFRDLTLTLKAQNK 420
NTEK KRP +EISF+DLTLTLKAQNK
Sbjct: 79 NTEKRKRPLMEISFKDLTLTLKAQNK 2
>TC218418 homologue to UP|Q6X4V5 (Q6X4V5) ABC transporter, partial (49%)
Length = 1220
Score = 186 bits (473), Expect = 3e-47
Identities = 104/256 (40%), Positives = 160/256 (61%), Gaps = 4/256 (1%)
Frame = +1
Query: 406 ISFRDLTLTLKAQN---KHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGK-ALGCL 461
++++DLT+ + N +++L +TG +PG TA+MGPSG+GK+T L AL+ + A
Sbjct: 241 LTWKDLTVMVTLSNGETQNVLEGLTGYAEPGTFTALMGPSGSGKSTLLDALSSRLAANAF 420
Query: 462 VTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLV 521
++G+IL+NGR + SF +V QDD + G LTV E + +SA+ RL ++ +K +
Sbjct: 421 LSGTILLNGRKAKL-SFGTA-AYVTQDDNLIGTLTVRETISYSARLRLPDNMPWADKRAL 594
Query: 522 VERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSAS 581
VE I +GLQ ++V+G RG+SGG+++RV++ LE++M P LL LDEPTSGLDSAS
Sbjct: 595 VESTIVAMGLQDCADTVIGNWHLRGISGGEKRRVSIALEILMRPRLLFLDEPTSGLDSAS 774
Query: 582 SQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSG 641
+ + + LR A +G + +HQPS +F +FD L LL G VY G A + E+F+
Sbjct: 775 AFFVTQTLRALARDGRTVIASIHQPSSEVFELFDQLYLL-SSGKTVYFGQASEAYEFFAQ 951
Query: 642 LGINVPERINPPDYYI 657
G P NP D+++
Sbjct: 952 AGFPCPALRNPSDHFL 999
>TC229551 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, partial (34%)
Length = 1436
Score = 166 bits (420), Expect = 5e-41
Identities = 100/256 (39%), Positives = 147/256 (57%), Gaps = 5/256 (1%)
Frame = +3
Query: 410 DLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLV-TGSILI 468
+ T+T K IL +TG +PGR+ AI+GPSG+GK+T L ALAG+ + TG ILI
Sbjct: 39 EATVTNGKNRKLILHGLTGYAQPGRLLAIIGPSGSGKSTLLDALAGRLTSNIKQTGKILI 218
Query: 469 NGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEF 528
NG + + G+V QDD + LT E L++SA + +S EK + +
Sbjct: 219 NGHKQELAYGTS--GYVTQDDAMLSCLTAGETLYYSAMLQFPNTMSVEEKKERADMTLRE 392
Query: 529 LGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLL-- 586
+GLQ N+ VG +G+SGGQR+R+++ +E++ P LL LDEPTSGLDSA+S ++
Sbjct: 393 MGLQDAINTRVGGWNCKGLSGGQRRRLSICIEILTHPKLLFLDEPTSGLDSAASYYVMSG 572
Query: 587 --RALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGI 644
++R+ ++ I VHQPS +F +F DL LL G VY G A ++F+ G
Sbjct: 573 IANLIQRDGIQR-TIVASVHQPSSEVFQLFHDLFLLSSGE-TVYFGPASDANQFFASNGF 746
Query: 645 NVPERINPPDYYIDIL 660
P NP D+Y+ I+
Sbjct: 747 PCPPLYNPSDHYLRII 794
>BE806563 similar to GP|11994752|db ABC transporter-like protein {Arabidopsis
thaliana}, partial (20%)
Length = 428
Score = 123 bits (309), Expect = 4e-28
Identities = 69/138 (50%), Positives = 90/138 (65%), Gaps = 2/138 (1%)
Frame = +1
Query: 479 KKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSV 538
K+ +GFVPQDDV + +LTV E L ++A RL LS+ EK E VI LGL RNS
Sbjct: 13 KRKVGFVPQDDVHYPHLTVLETLTYAALLRLPKSLSREEKKEHAEMVIAELGLTRCRNSP 192
Query: 539 VGTVEK--RGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEG 596
VG RG+SGG+RKRV++G EM++ PSLL +DEPTSGLDS ++QL++ L A G
Sbjct: 193 VGGCMALFRGISGGERKRVSIGQEMLVNPSLLFVDEPTSGLDSTTAQLIVSVLHGLARAG 372
Query: 597 VNICMVVHQPSYALFNMF 614
+ +HQPS L+ MF
Sbjct: 373 RTVVATIHQPSSRLYRMF 426
>TC222300 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {Arabidopsis
thaliana;} , partial (37%)
Length = 801
Score = 106 bits (264), Expect = 6e-23
Identities = 54/137 (39%), Positives = 83/137 (60%)
Frame = +1
Query: 521 VVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSA 580
+ + I +GLQ + ++G RG+SGG++KR+++ L ++ P LL LDEPTSGLDSA
Sbjct: 4 ITDATIIEMGLQDCDDRLIGKWHLRGISGGEKKRLSIAL*ILTRPRLLFLDEPTSGLDSA 183
Query: 581 SSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFS 640
S+ +++ LR A + + +HQP +F ++DDL LL GG VY G AK E+F
Sbjct: 184 SAFFVVQTLRNVARDERTVIYSIHQPDSEVFALYDDLFLL-SGGETVYFGEAKSAIEFFD 360
Query: 641 GLGINVPERINPPDYYI 657
G P + NP D+++
Sbjct: 361 EAGFPCPRKRNPYDHFL 411
>TC208020 similar to UP|Q76CU2 (Q76CU2) PDR-type ABC transporter 1, partial
(14%)
Length = 765
Score = 99.0 bits (245), Expect = 9e-21
Identities = 53/164 (32%), Positives = 94/164 (57%), Gaps = 6/164 (3%)
Frame = +2
Query: 520 LVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDS 579
+ +E V+E + L +RNS+VG G+S QRKR+ + +E+V PS++ +DEPTSGLD+
Sbjct: 158 MFIEEVMELVELNPLRNSLVGLPGVSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDA 337
Query: 580 ASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHG----SAKKV 635
++ +++R +R G + +HQPS +F FD+L L+ +GG +Y G + +
Sbjct: 338 RAAAIVMRTVRNTVDTGRTVVCTIHQPSIDIFEAFDELFLMKRGGQEIYVGPLGRHSTHL 517
Query: 636 EEYFSGLG--INVPERINPPDYYIDILEGIAAPGGSSGLSYQDL 677
+YF +G + + NP + +++ +A S G+ + DL
Sbjct: 518 IKYFESIGGVSKIKDGYNPATWMLEVT--TSAQELSLGVDFTDL 643
>CA820193 similar to PIR|T04229|T04 ABC-type transport protein F14M19.30 -
Arabidopsis thaliana, partial (23%)
Length = 423
Score = 94.0 bits (232), Expect = 3e-19
Identities = 46/117 (39%), Positives = 79/117 (67%), Gaps = 1/117 (0%)
Frame = +1
Query: 545 RGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREAL-EGVNICMVV 603
RG+SGG+R+RV++GL ++ +P++L+LDEPTSGLDS S+ ++R L++ + I + +
Sbjct: 70 RGLSGGERRRVSIGLCLLHDPAVLLLDEPTSGLDSTSAFKVMRILKQTCVSRNRTIILSI 249
Query: 604 HQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDIL 660
HQPS+ + D ++LL K G +V+HGS ++ + G VP ++N +Y ++IL
Sbjct: 250 HQPSFKILACIDRILLLSK-GQVVHHGSVATLQAFLHSNGFTVPHQLNALEYAMEIL 417
>BI701600
Length = 426
Score = 93.2 bits (230), Expect = 5e-19
Identities = 52/123 (42%), Positives = 75/123 (60%)
Frame = +3
Query: 418 QNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHS 477
+ + IL+ VTG +PG I A++GPSG+GK+T L ALAG+ G +TG+IL N +
Sbjct: 60 KERTILKGVTGIAQPGEILAVLGPSGSGKSTLLHALAGRLHGPGLTGTILANSSKLTKPV 239
Query: 478 FKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNS 537
++ GFV QDD+++ +LTV E L F A RL L + EK+ E I LGL N+
Sbjct: 240 LRR-TGFVTQDDILYPHLTVRETLVFCAMLRLPRALLRSEKLAAAEAAIAELGLGKCENT 416
Query: 538 VVG 540
++G
Sbjct: 417 IIG 425
>BI699796 similar to PIR|G85167|G85 ABC transporter like protein [imported] -
Arabidopsis thaliana, partial (13%)
Length = 421
Score = 89.7 bits (221), Expect = 6e-18
Identities = 44/125 (35%), Positives = 78/125 (62%)
Frame = +1
Query: 466 ILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERV 525
I I G + +F ++ G+ Q+D+ N+TVEE++ FSA RL + + K V V
Sbjct: 10 IRIGGYPKVQETFARVSGYCEQNDIHSPNITVEESVMFSAWLRLPSQIDAKTKAEFVNEV 189
Query: 526 IEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLL 585
I + L +++S+VG G+S QRKR+ + +E+V PS++ +DEPT+GLD+ ++ ++
Sbjct: 190 IHTIELDGIKDSLVGMPNISGLSTEQRKRLTIAVELVANPSIIFMDEPTTGLDARAAAVV 369
Query: 586 LRALR 590
+RA++
Sbjct: 370 MRAVK 384
>BQ080393
Length = 347
Score = 84.0 bits (206), Expect = 3e-16
Identities = 39/114 (34%), Positives = 74/114 (64%)
Frame = +2
Query: 462 VTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLV 521
+ G I I+G ++ +F ++ G+ Q D+ +T+ E+L +SA RL ++SK EK+
Sbjct: 5 IEGDIRISGFPKNQETFARVSGYCEQTDIHSPQVTIRESLLYSAYLRLPKEVSKDEKIQF 184
Query: 522 VERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTS 575
V++V++ + L ++++++VG G+S QRKR+ + +E+V PS++ +DEPTS
Sbjct: 185 VDQVMDLVELDNLKDAIVGLPGVTGLSTEQRKRLTIAVELVANPSIIFMDEPTS 346
>TC217803 similar to UP|Q9AT00 (Q9AT00) At1g65410/T8F5_19, partial (81%)
Length = 1563
Score = 75.1 bits (183), Expect = 1e-13
Identities = 76/277 (27%), Positives = 129/277 (46%), Gaps = 27/277 (9%)
Frame = +3
Query: 377 QQQENKNLTFSGVLK---MATNTEKSKRPFIEISFRDLTLTLKAQNKHILRNVTGKIKPG 433
+ Q++ + F+G K ++T + + I RD+ + K IL V+ KIK G
Sbjct: 246 KSQDSSAIHFNGSSKSEQLSTARDHEDDSDVLIECRDVYKSFG--EKKILNGVSFKIKHG 419
Query: 434 RITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGR-------NESIHSFKKIIGFVP 486
I+GPSG GK+T L +AG L G + I G+ ++ I + IG V
Sbjct: 420 EAVGIIGPSGTGKSTVLKIIAG--LLAPDKGEVYIRGKKRVGLVSDDDISGLR--IGLVF 587
Query: 487 QDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRG 546
Q + +LTV EN+ F S +S+ + +V + +GL+ V + +
Sbjct: 588 QSAALFDSLTVRENVGFLLYEHSS--MSEDQISELVTETLAAVGLKGVEDRLPSE----- 746
Query: 547 VSGGQRKRVNVGLEMV-------MEPSLLMLDEPTSGLDSASSQLLLRALRREALEG--- 596
+SGG +KRV + ++ +EP +L+ DEPT+GLD +S ++ +R ++G
Sbjct: 747 LSGGMKKRVALARSIICDTTKESIEPEVLLYDEPTAGLDPIASTVVEDLIRSVHIKGQDA 926
Query: 597 -------VNICMVVHQPSYALFNMFDDLILLGKGGLM 626
+ +V HQ S + D L+ L KG ++
Sbjct: 927 RGKPGNISSYVVVTHQHS-TIKRAIDRLLFLHKGKIV 1034
>TC218050 weakly similar to UP|Q7PC82 (Q7PC82) PDR14 ABC transporter, partial
(28%)
Length = 1436
Score = 73.6 bits (179), Expect = 4e-13
Identities = 35/108 (32%), Positives = 65/108 (59%), Gaps = 6/108 (5%)
Frame = +2
Query: 557 VGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDD 616
+ +E+V PS++ +DEPTSGLD+ ++ +++RA++ G +HQPS +F FD+
Sbjct: 2 IAVELVSNPSIIFMDEPTSGLDARAAAVVMRAVKNVVATGRTTVCTIHQPSIDIFETFDE 181
Query: 617 LILLGKGGLMVYHG----SAKKVEEYFSGL-GI-NVPERINPPDYYID 658
LIL+ GG ++Y G + ++ EYF + G+ + + NP + ++
Sbjct: 182 LILMKSGGRIIYSGMLGHHSSRLIEYFQNIPGVPKIKDNYNPATWMLE 325
>TC218748 similar to UP|Q76CU1 (Q76CU1) PDR-type ABC transporter 2
(Fragment), partial (37%)
Length = 1384
Score = 72.4 bits (176), Expect = 9e-13
Identities = 35/106 (33%), Positives = 64/106 (60%), Gaps = 6/106 (5%)
Frame = +3
Query: 560 EMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLIL 619
E+V PS++ +DEPTSGLD+ ++ +++R +R G + +HQPS +F FD+L+L
Sbjct: 9 ELVANPSIIFMDEPTSGLDARAAAIVMRTVRNTVDTGRTVVCTIHQPSIDIFESFDELLL 188
Query: 620 LGKGGLMVYHG----SAKKVEEYFSGL-GIN-VPERINPPDYYIDI 659
+ +GG +Y G + + YF G+ G+N + + NP + +++
Sbjct: 189 MKQGGQEIYVGPLGHHSSHLINYFEGIQGVNKIKDGYNPATWMLEV 326
>TC234765 similar to UP|Q84M24 (Q84M24) AtABCA1, partial (9%)
Length = 535
Score = 71.6 bits (174), Expect = 2e-12
Identities = 47/169 (27%), Positives = 91/169 (53%), Gaps = 2/169 (1%)
Frame = +3
Query: 425 NVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIH--SFKKII 482
++T ++ G +G +GAGKTT +S L G+ C G+ I G++ H + ++ I
Sbjct: 33 SLTFSVQEGECFGFLGTNGAGKTTTISMLCGEE--CPSDGTAFIFGKDICSHPKAARRYI 206
Query: 483 GFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTV 542
G+ PQ D + LTV E+L A+ + D + +V+E++ EF L+
Sbjct: 207 GYCPQFDALLEFLTVREHLELYARIKGVPDFAIDN--VVMEKLTEFDLLKHANKPSFS-- 374
Query: 543 EKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRR 591
+SGG +++++V + M+ +P +++LDEP++G+D + + + + R
Sbjct: 375 ----LSGGNKRKLSVAIAMIGDPPIVILDEPSTGMDPIAKRFMWDVISR 509
>TC219347 similar to UP|Q9LF78 (Q9LF78) Mitochondrial half-ABC transporter,
partial (36%)
Length = 1085
Score = 70.9 bits (172), Expect = 3e-12
Identities = 69/230 (30%), Positives = 116/230 (50%), Gaps = 12/230 (5%)
Frame = +1
Query: 406 ISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGS 465
I F ++ + + K IL ++ + G+ AI+G SG+GK+T L L +GS
Sbjct: 52 IQFENVHFSYLTERK-ILDGISFVVPAGKSVAIVGTSGSGKSTILRLLF--RFFDPHSGS 222
Query: 466 ILINGRN---ESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVV 522
I I+ +N ++ S +K IG VPQD V+ N T+ N+ + RLSA
Sbjct: 223 IKIDDQNIREVTLESLRKSIGVVPQDTVLF-NDTIFHNIHYG---RLSA---------TE 363
Query: 523 ERVIEFLGLQSVRNSVVGTVEK-------RGV--SGGQRKRVNVGLEMVMEPSLLMLDEP 573
E V E ++ N+++ +K RG+ SGG+++RV + + P++L+ DE
Sbjct: 364 EEVYEAAQQAAIHNTIMKFPDKYSTVVGERGLKLSGGEKQRVALARAFLKAPAILLCDEA 543
Query: 574 TSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKG 623
TS LDS + +L AL+ A +I + H+ + A+ D++I+L G
Sbjct: 544 TSALDSTTEAEILSALKSVANNRTSI-FIAHRLTTAM--QCDEIIVLENG 684
>TC207459 similar to UP|Q94IH6 (Q94IH6) CjMDR1, partial (30%)
Length = 1329
Score = 70.1 bits (170), Expect = 5e-12
Identities = 70/243 (28%), Positives = 127/243 (51%), Gaps = 8/243 (3%)
Frame = +1
Query: 401 RPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGC 460
RP I+I FRDL+LT+ + G+ A++G SG+GK+T ++ L +
Sbjct: 454 RPDIQI-FRDLSLTIHS---------------GKTVALVGESGSGKSTVIALL--QRFYN 579
Query: 461 LVTGSILING---RNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPE 517
+G I ++G R + ++ +G V Q+ V+ N T+ N+ + D ++ E
Sbjct: 580 PDSGQITLDGIEIRELQLKWLRQQMGLVSQEPVLF-NETIRANIAYGK----GGDATEAE 744
Query: 518 KVLVVE--RVIEFL-GLQSVRNSVVGTVEKRG--VSGGQRKRVNVGLEMVMEPSLLMLDE 572
+ E +F+ GLQ +++VG +RG +SGGQ++RV + ++ P +L+LDE
Sbjct: 745 IIAAAEMANAHKFISGLQQGYDTIVG---ERGTQLSGGQKQRVAIARAIIKSPKILLLDE 915
Query: 573 PTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSA 632
TS LD+ S +++ AL + + + +V H+ S + N D+I + K G++V G
Sbjct: 916 ATSALDAESERVVQDALDKVMVNRTTV-VVAHRLS-TIKNA--DVIAVVKNGVIVEKGKH 1083
Query: 633 KKV 635
+K+
Sbjct: 1084EKL 1092
>TC207563 similar to UP|Q9ZRG2 (Q9ZRG2) P-glycoprotein, partial (24%)
Length = 1164
Score = 67.8 bits (164), Expect = 2e-11
Identities = 53/182 (29%), Positives = 95/182 (52%), Gaps = 5/182 (2%)
Frame = +1
Query: 431 KPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILING---RNESIHSFKKIIGFVPQ 487
+ G+ A++GPSG GK++ ++ + + +G ++I+G R ++ S ++ I VPQ
Sbjct: 244 RAGKTLALVGPSGCGKSSIIALI--QRFYDPTSGRVMIDGKDIRKYNLKSLRRHISVVPQ 417
Query: 488 DDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGV 547
+ + T+ EN+ + + A++ + + + I GL + VG +RGV
Sbjct: 418 EPCLFAT-TIYENIAYGHESATEAEIIEAATLANAHKFIS--GLPDGYKTFVG---ERGV 579
Query: 548 --SGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQ 605
SGGQ++R+ V V + L++LDE TS LD+ S + + AL R A G +V H+
Sbjct: 580 QLSGGQKQRIAVARAFVRKAELMLLDEATSALDAESERSVQEALDR-ASSGKTTIIVAHR 756
Query: 606 PS 607
S
Sbjct: 757 LS 762
>TC213020 weakly similar to UP|Q8GU71 (Q8GU71) MDR-like ABC transporter,
partial (18%)
Length = 864
Score = 67.4 bits (163), Expect = 3e-11
Identities = 54/188 (28%), Positives = 96/188 (50%), Gaps = 5/188 (2%)
Frame = +1
Query: 425 NVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILING---RNESIHSFKKI 481
N+ ++ G+ A++G SG+GK+T +S + +G +L++ +N ++ S +
Sbjct: 4 NLNLRVPAGKSLAVVGQSGSGKSTVISLVM--RFYDPDSGLVLVDECDIKNLNLRSLRLR 177
Query: 482 IGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGT 541
IG V Q+ + TV EN+ + + ++ K K I +
Sbjct: 178 IGLVQQEPALFST-TVYENIKYGKEEASEIEVMKAAKAANAHEFIS-----RMPEGYKTE 339
Query: 542 VEKRGV--SGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNI 599
V +RGV SGGQ++RV + ++ +PS+L+LDE TS LD+ S +L+ AL + +EG
Sbjct: 340 VGERGVQLSGGQKQRVAIARAILKDPSILLLDEATSALDTVSERLVQEALDK-LMEGRTT 516
Query: 600 CMVVHQPS 607
+V H+ S
Sbjct: 517 ILVAHRLS 540
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,357,851
Number of Sequences: 63676
Number of extensions: 692060
Number of successful extensions: 4588
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 4512
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4567
length of query: 1007
length of database: 12,639,632
effective HSP length: 107
effective length of query: 900
effective length of database: 5,826,300
effective search space: 5243670000
effective search space used: 5243670000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144720.7


