

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144720.2 + phase: 0 /pseudo
(552 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
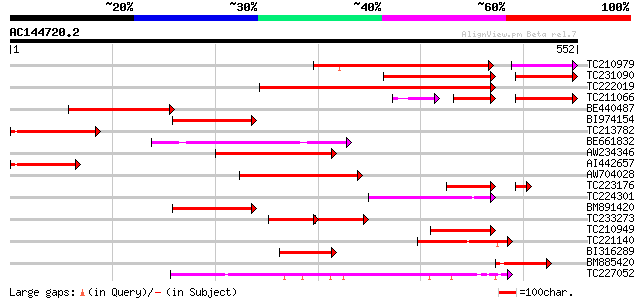
Score E
Sequences producing significant alignments: (bits) Value
TC210979 244 2e-75
TC231090 195 3e-71
TC222019 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-li... 251 6e-67
TC211066 92 1e-41
BE440487 167 1e-41
BI974154 153 2e-37
TC213782 similar to UP|Q6YU96 (Q6YU96) Amino acid transporter-li... 145 6e-35
BE661832 138 7e-33
AW234346 119 4e-27
AI442657 117 2e-26
AW704028 108 5e-24
TC223176 weakly similar to UP|Q8LPF4 (Q8LPF4) AT5g02170/T7H20_22... 89 3e-22
TC224301 weakly similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, par... 96 4e-20
BM891420 similar to GP|20197120|gb| expressed protein {Arabidops... 92 5e-19
TC233273 91 2e-18
TC210949 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-li... 73 3e-13
TC221140 similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, partial (20%) 73 4e-13
BI316289 69 5e-12
BM885420 65 6e-11
TC227052 weakly similar to GB|AAM19768.1|20453045|AY094388 AT3g5... 64 2e-10
>TC210979
Length = 928
Score = 244 bits (623), Expect(2) = 2e-75
Identities = 120/179 (67%), Positives = 142/179 (79%), Gaps = 3/179 (1%)
Frame = +2
Query: 296 LSYISAGGVIASVLVVLCLLWIGI---EDVGFQRSGTTLNLGTLPVAIGLYGYCYSGHAV 352
LSYISA GVIA++LVVLCL W+G+ D+ Q + T NL T PVAIGLYGYCY+GHAV
Sbjct: 2 LSYISACGVIATILVVLCLFWVGLLDNADIHTQGTTKTFNLATFPVAIGLYGYCYAGHAV 181
Query: 353 FPNIYTSMAKPNQFPAVLVACFGVCTLLYAGGAVMGYKMFGEDTLSQFTLNLPQDLVATK 412
F NIYT+MA NQFP VL+ CF +CT +Y A+MGY FG+ TLSQ+TLN+PQ LVATK
Sbjct: 182 FSNIYTAMANRNQFPGVLLVCFAICTSMYCAVAIMGYTAFGKATLSQYTLNMPQHLVATK 361
Query: 413 IAVWTTVVNPFTKYALTISPVAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLSVP 471
IAVWTTVVNPFTKYAL++SPVAM LEELIPAN +++S IRT LV STL++GLSVP
Sbjct: 362 IAVWTTVVNPFTKYALSLSPVAMCLEELIPANSPNFFIYSKLIRTALVVSTLLVGLSVP 538
Score = 57.4 bits (137), Expect(2) = 2e-75
Identities = 27/64 (42%), Positives = 38/64 (59%)
Frame = +1
Query: 489 LCRFGDVLDWIITYNACNFDIAMCLLPKNLAGKSNANTGRTLHYNHCSRRCVFFRWNVLS 548
L RFGDV++WI +N C D + CL K+ +++A +G L YN+CSR C +LS
Sbjct: 538 LXRFGDVINWIFAHNVCVLDSSRCLFLKHKGWQNHALSGINLCYNYCSRHCFKLFGIILS 717
Query: 549 PCGD 552
P D
Sbjct: 718 PLRD 729
>TC231090
Length = 559
Score = 195 bits (496), Expect(2) = 3e-71
Identities = 95/109 (87%), Positives = 103/109 (94%)
Frame = +2
Query: 365 QFPAVLVACFGVCTLLYAGGAVMGYKMFGEDTLSQFTLNLPQDLVATKIAVWTTVVNPFT 424
QFP VL+ACFG+CTLLYAG AV+GY MFGE LSQFTLN+P++LVATKIAVWTTVVNPFT
Sbjct: 2 QFPGVLLACFGICTLLYAGAAVLGYTMFGEAILSQFTLNMPKELVATKIAVWTTVVNPFT 181
Query: 425 KYALTISPVAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLSVPFF 473
KYALTISPVAMSLEELIP+NHAKSYL+SIFIRTGLV STLVIGLSVPFF
Sbjct: 182 KYALTISPVAMSLEELIPSNHAKSYLYSIFIRTGLVLSTLVIGLSVPFF 328
Score = 92.0 bits (227), Expect(2) = 3e-71
Identities = 39/60 (65%), Positives = 47/60 (78%)
Frame = +1
Query: 493 GDVLDWIITYNACNFDIAMCLLPKNLAGKSNANTGRTLHYNHCSRRCVFFRWNVLSPCGD 552
GDV+DWIIT+NAC FD +CL PKNLAG+S+ NTG TLHYN+ SR C+F W +L PC D
Sbjct: 334 GDVIDWIITHNACYFDTTLCLFPKNLAGQSDTNTGSTLHYNYHSRCCMFCFWIILRPCRD 513
>TC222019 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-like, partial
(84%)
Length = 979
Score = 251 bits (641), Expect = 6e-67
Identities = 117/231 (50%), Positives = 166/231 (71%), Gaps = 1/231 (0%)
Frame = +1
Query: 244 IEYIILEGDNLASLFPNAYLNLGGIELNPQTLFAVVAALAVLPTVWLRDLSVLSYISAGG 303
+E+I LEGDNL LFP L+LG L+ LF ++AAL ++PTVWL+DL ++S +SAGG
Sbjct: 19 LEFITLEGDNLTGLFPGTSLDLGSFRLDSVHLFGILAALIIIPTVWLKDLRIISILSAGG 198
Query: 304 VIASVLVVLCLLWIG-IEDVGFQRSGTTLNLGTLPVAIGLYGYCYSGHAVFPNIYTSMAK 362
V A++L+V+C+ +G I VGF +G +N +P+AIG++G+C++GH+VFPNIY SMA
Sbjct: 199 VFATLLIVVCVFCVGTINGVGFHHTGQLVNWSGIPLAIGIHGFCFAGHSVFPNIYQSMAD 378
Query: 363 PNQFPAVLVACFGVCTLLYAGGAVMGYKMFGEDTLSQFTLNLPQDLVATKIAVWTTVVNP 422
QF L+ CF + +Y G A+MG+ MFG +TLSQ TLN+P+D A+K+A+WTTV+NP
Sbjct: 379 KRQFTKALIICFVLSITIYGGVAIMGFLMFGGETLSQITLNMPRDAFASKVALWTTVINP 558
Query: 423 FTKYALTISPVAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLSVPFF 473
FTKYAL ++P+A SLEEL+P + +Y I +RT LV ST+ +PFF
Sbjct: 559 FTKYALLMNPLARSLEELLPDRISSTYRCFILLRTALVVSTVCAAFLIPFF 711
>TC211066
Length = 805
Score = 91.7 bits (226), Expect(3) = 1e-41
Identities = 39/60 (65%), Positives = 47/60 (78%)
Frame = +1
Query: 493 GDVLDWIITYNACNFDIAMCLLPKNLAGKSNANTGRTLHYNHCSRRCVFFRWNVLSPCGD 552
GDV+DWIIT+NAC FD +CL PKNLAG+S+ NTG TLHYN+ SR C+F WN+L P D
Sbjct: 244 GDVIDWIITHNACYFDTTLCLFPKNLAGQSDTNTGSTLHYNYHSRCCMFCFWNILRPLRD 423
Score = 75.1 bits (183), Expect(3) = 1e-41
Identities = 37/41 (90%), Positives = 39/41 (94%)
Frame = +2
Query: 433 VAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLSVPFF 473
VAMSLEELIP+NHAKSYL+SIFIRTGLV STL IGLSVPFF
Sbjct: 116 VAMSLEELIPSNHAKSYLYSIFIRTGLVLSTLFIGLSVPFF 238
Score = 42.7 bits (99), Expect(3) = 1e-41
Identities = 23/46 (50%), Positives = 27/46 (58%)
Frame = +3
Query: 373 CFGVCTLLYAGGAVMGYKMFGEDTLSQFTLNLPQDLVATKIAVWTT 418
C+G C G E LSQFTLN+P++LVAT IAVWTT
Sbjct: 6 CWGCCN---------GIHNV*EAILSQFTLNMPKELVATNIAVWTT 116
>BE440487
Length = 414
Score = 167 bits (422), Expect = 1e-41
Identities = 88/106 (83%), Positives = 98/106 (92%), Gaps = 3/106 (2%)
Frame = -3
Query: 58 YRQSIDLYSSVPSPSIGFLGNSSLTRLSSSFLSTSLTRRHTPEVLPSVTKPLI-QPTEEE 116
+RQSIDLYSSVPSP+IGFLG SL+RLSSSFLSTSLTRRHTPE LPS+TKPLI Q TE+E
Sbjct: 331 FRQSIDLYSSVPSPNIGFLGTPSLSRLSSSFLSTSLTRRHTPEALPSLTKPLIQQDTEDE 152
Query: 117 KHQRRSSHTLLPPL-SRRSSLLKKESKVSH-EVPSRHCSFGQAVLN 160
+HQRRSSHTLLPPL SRRSSL+KK+SKV+H EVPSRHCSFGQ +LN
Sbjct: 151 QHQRRSSHTLLPPLPSRRSSLIKKDSKVAHLEVPSRHCSFGQDMLN 14
>BI974154
Length = 420
Score = 153 bits (387), Expect = 2e-37
Identities = 76/82 (92%), Positives = 77/82 (93%)
Frame = +2
Query: 159 LNGINVLCGVGILSTPYAAKEGGWLGLSILFIFGILSFYTGLLLRSCLDSEPGLETYPDI 218
LNGINVLCGVGILSTPYAAK GGWLGLSIL IF I+SFYTGLLLRSCLDSEP LETYPDI
Sbjct: 2 LNGINVLCGVGILSTPYAAKVGGWLGLSILVIFAIISFYTGLLLRSCLDSEPELETYPDI 181
Query: 219 GQAAFGTAGRIAISIVLYVELY 240
GQAAFGT GRIAISIVLYVELY
Sbjct: 182 GQAAFGTTGRIAISIVLYVELY 247
>TC213782 similar to UP|Q6YU96 (Q6YU96) Amino acid transporter-like protein,
partial (15%)
Length = 435
Score = 145 bits (365), Expect = 6e-35
Identities = 77/90 (85%), Positives = 80/90 (88%), Gaps = 2/90 (2%)
Frame = +1
Query: 1 MNNSVASENSFIIESDEEDD-KDFNKGD-DGNDSDSSNYSNENPPQRKQSSYNPSWPQSY 58
MNNSV SENSFIIESDEED+ KD NKG DGNDSDSSNYSNENPPQRK SSYN SWPQSY
Sbjct: 169 MNNSV-SENSFIIESDEEDEEKDLNKGGVDGNDSDSSNYSNENPPQRKPSSYNISWPQSY 345
Query: 59 RQSIDLYSSVPSPSIGFLGNSSLTRLSSSF 88
RQSIDLYSSVPSP+IG+LG SL RLSSSF
Sbjct: 346 RQSIDLYSSVPSPNIGYLGTPSLXRLSSSF 435
>BE661832
Length = 635
Score = 138 bits (347), Expect = 7e-33
Identities = 72/195 (36%), Positives = 110/195 (55%), Gaps = 1/195 (0%)
Frame = +1
Query: 139 KESKVSHEVPSRHCSFGQAVLNGINVLCGVGILSTPYAAKEGGWLGLSILFIFGILSFYT 198
+ +SH SF NGIN + S PY GGWL L++LF +FYT
Sbjct: 79 RREPLSHPSTENTASFFHTCFNGINAV------SVPYTLASGGWLSLALLFAIAAATFYT 240
Query: 199 GLLLRSCLDSEPGLETYPDIGQAAFGTAGRIAISIVLYVELYGCCIEYIILEGDNLASLF 258
G+L++ +D + + TYPD+G+ AFG GR+ IS ++Y EL+ + ++ILEGDNL++LF
Sbjct: 241 GVLIKR*MDKDSNIRTYPDMGELAFGKTGRLIISGLIYTELFLVSVGFLILEGDNLSNLF 420
Query: 259 PNAYLNLGGIELNPQTLFAVVAALAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIG 318
P ++ + + + LF ++ AL L +L +LSY+SA V AS +++L + W
Sbjct: 421 PTVEIHTADLAIGGKKLFVILVALV------LDNLRILSYVSASRVFASAIIILSISWTA 582
Query: 319 IED-VGFQRSGTTLN 332
D VGF + GT N
Sbjct: 583 TFDGVGFHQXGTLGN 627
>AW234346
Length = 371
Score = 119 bits (298), Expect = 4e-27
Identities = 56/118 (47%), Positives = 80/118 (67%)
Frame = +2
Query: 201 LLRSCLDSEPGLETYPDIGQAAFGTAGRIAISIVLYVELYGCCIEYIILEGDNLASLFPN 260
L++ C+D +P ++ +PDIGQ AFG GRI +SI + EL+ ++ILEGDNL L PN
Sbjct: 11 LVKRCMDMDPDIKNFPDIGQRAFGDKGRIIVSIAMNSELFLVVTGFLILEGDNLNKLVPN 190
Query: 261 AYLNLGGIELNPQTLFAVVAALAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIG 318
L L G+ + T+F ++AAL +LPTV L DLS+LSY+SA G +AS + +L + W G
Sbjct: 191 MQLELAGLTIGGTTIFTMIAALVILPTVLLEDLSMLSYVSASGALASSIFLLSIYWNG 364
>AI442657
Length = 381
Score = 117 bits (292), Expect = 2e-26
Identities = 62/71 (87%), Positives = 64/71 (89%), Gaps = 2/71 (2%)
Frame = +2
Query: 1 MNNSVASENSFIIESDEEDD-KDFNKGDDG-NDSDSSNYSNENPPQRKQSSYNPSWPQSY 58
MNNSV SENSFIIESDEED+ KD NKG+ G NDSDSSNYSNENPPQRK SSYN SWPQSY
Sbjct: 170 MNNSV-SENSFIIESDEEDEEKDLNKGEGGGNDSDSSNYSNENPPQRKPSSYNISWPQSY 346
Query: 59 RQSIDLYSSVP 69
RQSIDLYSSVP
Sbjct: 347 RQSIDLYSSVP 379
>AW704028
Length = 373
Score = 108 bits (271), Expect = 5e-24
Identities = 55/121 (45%), Positives = 78/121 (64%), Gaps = 1/121 (0%)
Frame = +3
Query: 224 GTAGRIAISIVLYVELYGCCIEYIILEGDNLASLFPNAYLNLGGIELNPQTLFAVVAALA 283
G GR+ IS +Y ELY I ++ILEGDNL +L P + + G + + LF ++ AL
Sbjct: 6 GKIGRLIISGYMYTELYLVSIGFLILEGDNLNNLCPIEEVQIAGFVIGGKQLFVILVALI 185
Query: 284 VLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIGIED-VGFQRSGTTLNLGTLPVAIGL 342
+LPTVWL +LS+LSY+SA GV AS +++L + W G D VGF + GT +N +P A+ L
Sbjct: 186 ILPTVWLDNLSMLSYVSASGVFASAVIILSISWTGTFDGVGFHQKGTLVNWRGIPTAVSL 365
Query: 343 Y 343
Y
Sbjct: 366 Y 368
>TC223176 weakly similar to UP|Q8LPF4 (Q8LPF4) AT5g02170/T7H20_220, partial
(11%)
Length = 437
Score = 88.6 bits (218), Expect(2) = 3e-22
Identities = 44/48 (91%), Positives = 46/48 (95%)
Frame = +2
Query: 426 YALTISPVAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLSVPFF 473
YALTISPVAMSLEELIP+NHAKSYL+SIFIRTGLV STL IGLSVPFF
Sbjct: 233 YALTISPVAMSLEELIPSNHAKSYLYSIFIRTGLVLSTLFIGLSVPFF 376
Score = 35.0 bits (79), Expect(2) = 3e-22
Identities = 13/16 (81%), Positives = 15/16 (93%)
Frame = +1
Query: 493 GDVLDWIITYNACNFD 508
GDV+DWIIT+NAC FD
Sbjct: 382 GDVIDWIITHNACYFD 429
>TC224301 weakly similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, partial (29%)
Length = 667
Score = 95.9 bits (237), Expect = 4e-20
Identities = 46/124 (37%), Positives = 74/124 (59%)
Frame = +2
Query: 350 HAVFPNIYTSMAKPNQFPAVLVACFGVCTLLYAGGAVMGYKMFGEDTLSQFTLNLPQDLV 409
H + Y SM +QF CF VC L YA V+ Y MFG++ SQ TLNLP
Sbjct: 2 HPIXXTXYNSMRDKSQFSRXXSICFSVCXLGYAAAGVLXYLMFGQEVESQVTLNLPTGKF 181
Query: 410 ATKIAVWTTVVNPFTKYALTISPVAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLS 469
++ +A++TT+VNP TKYAL ++PV +++ + ++ K +F+ T ++ STL++ ++
Sbjct: 182 SSHVAIFTTLVNPITKYALMLTPVIYAVKNKVSWHYNKRST-HMFVITSMLISTLIVAVA 358
Query: 470 VPFF 473
+P F
Sbjct: 359 IPLF 370
>BM891420 similar to GP|20197120|gb| expressed protein {Arabidopsis
thaliana}, partial (12%)
Length = 433
Score = 92.4 bits (228), Expect = 5e-19
Identities = 39/82 (47%), Positives = 57/82 (68%)
Frame = +2
Query: 159 LNGINVLCGVGILSTPYAAKEGGWLGLSILFIFGILSFYTGLLLRSCLDSEPGLETYPDI 218
L+ NV+ GVGILSTPY KE GW+ + ++ +F ++ YT L+R C +S G+ +YPDI
Sbjct: 164 LSATNVMAGVGILSTPYTLKEAGWMSMVLMVLFAVICCYTATLMRYCFESREGITSYPDI 343
Query: 219 GQAAFGTAGRIAISIVLYVELY 240
G+AAFG GRI +S+ L + +Y
Sbjct: 344 GEAAFGKYGRIIVSVSLSIFIY 409
>TC233273
Length = 601
Score = 90.5 bits (223), Expect = 2e-18
Identities = 39/52 (75%), Positives = 45/52 (86%)
Frame = +1
Query: 298 YISAGGVIASVLVVLCLLWIGIEDVGFQRSGTTLNLGTLPVAIGLYGYCYSG 349
++ AGGV+AS+LVV CLLW+GIEDVGF GTTLNL TLPV +GLYGYCYSG
Sbjct: 445 FVIAGGVVASILVVXCLLWVGIEDVGFHSKGTTLNLATLPVDVGLYGYCYSG 600
Score = 80.1 bits (196), Expect = 2e-15
Identities = 38/48 (79%), Positives = 44/48 (91%)
Frame = +1
Query: 253 NLASLFPNAYLNLGGIELNPQTLFAVVAALAVLPTVWLRDLSVLSYIS 300
+L+SLFP+A+LNLGGIELN TLFAV+ LAVLPTVWLRDLS+LSYIS
Sbjct: 4 SLSSLFPSAHLNLGGIELNSHTLFAVITTLAVLPTVWLRDLSILSYIS 147
>TC210949 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-like, partial
(35%)
Length = 569
Score = 73.2 bits (178), Expect = 3e-13
Identities = 34/64 (53%), Positives = 47/64 (73%)
Frame = +2
Query: 410 ATKIAVWTTVVNPFTKYALTISPVAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLS 469
A+K+A+WTTV+NP TKYAL ++P+A SLEEL+P + SY + +RT LV ST+ +
Sbjct: 11 ASKVALWTTVINPLTKYALLMNPLARSLEELLPDRISSSYWCFMLLRTTLVASTVCVAFL 190
Query: 470 VPFF 473
VPFF
Sbjct: 191 VPFF 202
>TC221140 similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, partial (20%)
Length = 644
Score = 72.8 bits (177), Expect = 4e-13
Identities = 39/95 (41%), Positives = 65/95 (68%), Gaps = 3/95 (3%)
Frame = +1
Query: 398 SQFTLNLPQDLVATKIAVWTTVVNPFTKYALTISPVAMSLEELIPANHAKSYLFSIFIRT 457
SQ TLNLP + V++K+A++TT+VNP +K+AL +P+ +L++L+P + K+ +I + T
Sbjct: 10 SQVTLNLPLNKVSSKLAIYTTLVNPISKFALMATPITNALKDLLPRAY-KNRATNILVST 186
Query: 458 GLVFSTLVIGLSVPFF---VALCYGKLAITCLIIL 489
LV S ++ LSVPFF ++L L++T I+L
Sbjct: 187 VLVISATIVALSVPFFGDLMSLVGAFLSVTASILL 291
>BI316289
Length = 412
Score = 68.9 bits (167), Expect = 5e-12
Identities = 35/56 (62%), Positives = 42/56 (74%)
Frame = +3
Query: 263 LNLGGIELNPQTLFAVVAALAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIG 318
LNLGGIELN +TLFAV+ LAVLPTVWLRDLS+LSYIS + +V+ L +G
Sbjct: 3 LNLGGIELNSRTLFAVITTLAVLPTVWLRDLSILSYISGKSLFLVQVVISYPLKLG 170
>BM885420
Length = 421
Score = 65.5 bits (158), Expect = 6e-11
Identities = 30/54 (55%), Positives = 37/54 (67%)
Frame = +3
Query: 474 VALCYGKLAITCLIILCRFGDVLDWIITYNACNFDIAMCLLPKNLAGKSNANTG 527
V LC L + + R GDV+DWIIT+NAC FD +CL PKNLAG+S+ NTG
Sbjct: 180 VTLCI--LTANIIFLFYRSGDVIDWIITHNACYFDTTLCLFPKNLAGQSDTNTG 335
>TC227052 weakly similar to GB|AAM19768.1|20453045|AY094388
AT3g56200/F18O21_160 {Arabidopsis thaliana;} , partial
(68%)
Length = 1627
Score = 63.5 bits (153), Expect = 2e-10
Identities = 87/369 (23%), Positives = 151/369 (40%), Gaps = 36/369 (9%)
Frame = +3
Query: 157 AVLNGINVLCGVGILSTPYAAKEGGWL-GLSILFIFGILS-FYTGLLLRSCLDSEPGLET 214
AV N + G GI+S P K G + +++ + +L+ L+R E T
Sbjct: 156 AVFNVATSIVGAGIMSIPAIMKVLGVVPAFAMILVVAVLAELSVDFLMRFTHSGET--TT 329
Query: 215 YPDIGQAAFGTAGRIAISIVLYVELYGCCIEYIILEGDNLASLFPNAYLNLG------GI 268
Y + + AFG+ G +A + + + G I Y+I+ GD L+ ++LG GI
Sbjct: 330 YAGVMREAFGSGGALAAQVCVIITNVGGLILYLIIIGDVLSGKQNGGEVHLGILQQWFGI 509
Query: 269 ELNPQTLFAVVAALA--VLPTVWLRDLSVLSYISAGGVIASVLVV--LCLLWIGIEDVG- 323
FA++ L +LP V + + L Y SA + +V V C L I G
Sbjct: 510 HWWNSREFALLFTLVFVMLPLVLYKRVESLKYSSAVSTLLAVAFVGICCGLAITALVQGK 689
Query: 324 ------FQRSGTTLNLGTLPVAIGLYGYCYSGHAVFPNIYTSMAKPNQFPAVLVACFGVC 377
F R + L A+ + ++ H I +AK +Q + +C
Sbjct: 690 TQTPRLFPRLDYQTSFFDLFTAVPVVVTAFTFHFNVHPIGFELAKASQMTTAVRLALLLC 869
Query: 378 TLLYAGGAVMGYKMFGEDTLSQFTLNLPQD-----------LVATKIAVWTTVVNPFTKY 426
++Y + GY +FG+ T S +N Q+ LV A+ +V P +
Sbjct: 870 AVIYLAIGLFGYMLFGDSTQSDILINFDQNAGSAVGSLLNSLVRVSYALHIMLVFPLLNF 1049
Query: 427 AL--TISPVAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLSVP----FFVALCYGK 480
+L I V + ++ ++ + + ++ + LVFS L +++P FF L G
Sbjct: 1050SLRTNIDEVLFPKKPMLATDNKRFMILTLVL---LVFSYLA-AIAIPDIWYFFQFL--GS 1211
Query: 481 LAITCLIIL 489
+ CL +
Sbjct: 1212SSAVCLAFI 1238
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,626,291
Number of Sequences: 63676
Number of extensions: 471596
Number of successful extensions: 3011
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 2925
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2983
length of query: 552
length of database: 12,639,632
effective HSP length: 102
effective length of query: 450
effective length of database: 6,144,680
effective search space: 2765106000
effective search space used: 2765106000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144720.2


