

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.4 + phase: 0
(776 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
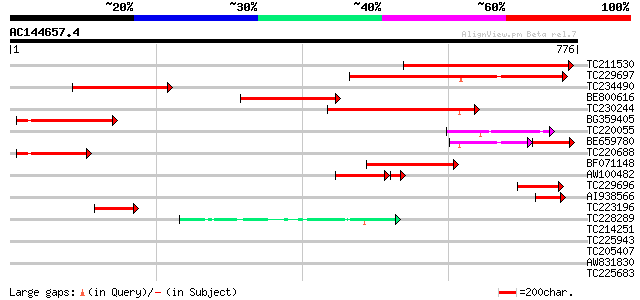
Score E
Sequences producing significant alignments: (bits) Value
TC211530 weakly similar to UP|Q8LPA2 (Q8LPA2) Chloride channel, ... 363 e-100
TC229697 similar to UP|CLCB_ARATH (P92942) Chloride channel prot... 290 2e-78
TC234490 weakly similar to UP|Q8LPA2 (Q8LPA2) Chloride channel, ... 237 2e-62
BE800616 weakly similar to GP|21321024|db chloride channel {Oryz... 236 4e-62
TC230244 similar to UP|CLCB_ARATH (P92942) Chloride channel prot... 221 9e-58
BG359405 similar to GP|4768916|gb| CLC-Nt2 protein {Nicotiana ta... 124 2e-28
TC220055 homologue to GB|AAC26247.2|6382041|F9D12 Arabidopsis th... 112 5e-25
BE659780 similar to GP|4768916|gb| CLC-Nt2 protein {Nicotiana ta... 80 2e-24
TC220688 similar to UP|Q9XF71 (Q9XF71) CLC-Nt2 protein, partial ... 105 6e-23
BF071148 weakly similar to GP|1742959|emb CLC-d chloride channel... 89 1e-17
AW100482 similar to PIR|T52107|T52 anion channel protein CLC-a [... 76 7e-15
TC229696 similar to UP|Q9XF71 (Q9XF71) CLC-Nt2 protein, partial ... 76 5e-14
AI938566 66 7e-11
TC223196 similar to UP|Q9XF71 (Q9XF71) CLC-Nt2 protein, partial ... 64 3e-10
TC228289 48 1e-05
TC214251 33 0.62
TC225943 similar to UP|NU4C_LOTJA (Q9BBP3) NAD(P)H-quinone oxido... 31 2.4
TC205407 similar to UP|Q9XHD3 (Q9XHD3) PHAP2B protein, partial (... 30 3.1
AW831830 similar to GP|15912249|gb| AT5g54590/MRB17_9 {Arabidops... 30 3.1
TC225683 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit ... 30 5.3
>TC211530 weakly similar to UP|Q8LPA2 (Q8LPA2) Chloride channel, partial
(24%)
Length = 897
Score = 363 bits (932), Expect = e-100
Identities = 181/233 (77%), Positives = 205/233 (87%), Gaps = 1/233 (0%)
Frame = +2
Query: 540 VSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIV 599
+SK+VA+ FNAN+YD+IMKAKGLPYLETHAEPYMRQL+VGDVVTGPLQ FNG+EKV NIV
Sbjct: 11 ISKTVADAFNANIYDIIMKAKGLPYLETHAEPYMRQLSVGDVVTGPLQTFNGVEKVCNIV 190
Query: 600 FILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSS 659
FILRTT HNGFPVIDEPP S+AP+LFGIILR HL TLLKKKAF+ SP+A S DV+ +FS+
Sbjct: 191 FILRTTGHNGFPVIDEPPISQAPVLFGIILRDHLLTLLKKKAFMSSPMATSGDVINEFSA 370
Query: 660 DDFAKKYSVE-RVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGL 718
DDFAKK S + R+KIEDIQL+EEEM MF+DLHPFTNASPYTVVETMSL KAL LFRE+GL
Sbjct: 371 DDFAKKGSSKGRLKIEDIQLSEEEMDMFIDLHPFTNASPYTVVETMSLGKALTLFRELGL 550
Query: 719 RHLLVIPKIPGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLRFWQTFLEK 771
RHLLV+PK GRSPVVGILTRHDF EHILG+HPFLV++ K LRF +L K
Sbjct: 551 RHLLVVPKFSGRSPVVGILTRHDFMSEHILGLHPFLVRNTGKSLRF*HPWLGK 709
>TC229697 similar to UP|CLCB_ARATH (P92942) Chloride channel protein CLC-b
(AtCLC-b), partial (39%)
Length = 988
Score = 290 bits (741), Expect = 2e-78
Identities = 157/310 (50%), Positives = 217/310 (69%), Gaps = 12/310 (3%)
Frame = +1
Query: 466 AGLFVPIIVTGASYGRLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELT 525
+GLF+PII+ G+ YGRL+GI +G TN+ GL+AVLGAASL+ GSMR TVSLCVI LELT
Sbjct: 7 SGLFLPIILMGSGYGRLLGIYMGPHTNIDQGLFAVLGAASLMAGSMRMTVSLCVIFLELT 186
Query: 526 NNLLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDV--VT 583
NNLLLLP+ M+VL+++K+V + FN ++Y++I+ KGLP+++ + EP+MR LTVG++ V
Sbjct: 187 NNLLLLPITMIVLLIAKTVGDSFNPSIYEIILHLKGLPFMDANPEPWMRNLTVGELVDVK 366
Query: 584 GPLQMFNGIEKVRNIVFILRTTAHNGFPVIDE---PP----GSEAPILFGIILRHHLTTL 636
+ +G+EKV IV +L+ T HN FPV+D+ PP + L G+ILR HL
Sbjct: 367 PSVVTLHGVEKVAKIVDVLKNTTHNAFPVMDDGVVPPVVGQANGGTELHGLILRAHLIQA 546
Query: 637 LKKKAFLPS-PVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNA 695
LKKK FL ++V KF+ + A++ IE++ +T EEM MFVDLHP TN
Sbjct: 547 LKKKWFLKERRRTEEWEVREKFTWVELAEREG----SIEEVAVTSEEMEMFVDLHPLTNT 714
Query: 696 SPYTVVETMSLAKALILFREVGLRHLLVIPK--IPGRSPVVGILTRHDFTPEHILGMHPF 753
+P+TV+E+MS+AKA+ LFR+VGLRHLLV+PK G SPV+GILTR D ++IL + P
Sbjct: 715 TPFTVLESMSVAKAMTLFRQVGLRHLLVVPKYQASGISPVIGILTRQDLLAQNILTVFPH 894
Query: 754 LVKSRWKRLR 763
L KS+ + R
Sbjct: 895 LAKSKGREKR 924
>TC234490 weakly similar to UP|Q8LPA2 (Q8LPA2) Chloride channel, partial
(16%)
Length = 419
Score = 237 bits (604), Expect = 2e-62
Identities = 116/137 (84%), Positives = 124/137 (89%)
Frame = +2
Query: 87 GFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLFASIITAFIAPTA 146
G +GFCNNLAVENLAGIKFV TSNMMLERRF+ AF +FF SNL LT+FA ITA IAPTA
Sbjct: 8 GLVGFCNNLAVENLAGIKFVVTSNMMLERRFLMAFLVFFVSNLVLTVFACTITALIAPTA 187
Query: 147 AGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALL 206
GSGI EVKAYLNGVDAPGIFTVRTL VKIIGSITAVS SL+IGKAGPMVHTGACVAALL
Sbjct: 188 TGSGIPEVKAYLNGVDAPGIFTVRTLLVKIIGSITAVSSSLLIGKAGPMVHTGACVAALL 367
Query: 207 GQGGSKRYGITWRWLRF 223
GQGGSKRYG+TW+WL+F
Sbjct: 368 GQGGSKRYGLTWKWLKF 418
>BE800616 weakly similar to GP|21321024|db chloride channel {Oryza sativa
(japonica cultivar-group)}, partial (16%)
Length = 413
Score = 236 bits (601), Expect = 4e-62
Identities = 108/136 (79%), Positives = 119/136 (87%)
Frame = +1
Query: 317 LVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFG 376
LVDVPPVF+L V+GG+LGSLFN + +KVLRIYN INEKGTI ++ LACLISIFTSCLLFG
Sbjct: 4 LVDVPPVFVLGVIGGILGSLFNLILSKVLRIYNFINEKGTIFKILLACLISIFTSCLLFG 183
Query: 377 LPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHT 436
LPWL CRPCPPD EPCPTIGRSGIYKKFQCPPNHYN LASLIFNTNDDAIRNLFS +T
Sbjct: 184 LPWLTSCRPCPPDPSEPCPTIGRSGIYKKFQCPPNHYNDLASLIFNTNDDAIRNLFSKNT 363
Query: 437 DNEFELSSMLVFFIIC 452
D+EFE S+ + FI C
Sbjct: 364 DDEFEFKSVFIXFITC 411
>TC230244 similar to UP|CLCB_ARATH (P92942) Chloride channel protein CLC-b
(AtCLC-b), partial (30%)
Length = 709
Score = 221 bits (563), Expect = 9e-58
Identities = 114/217 (52%), Positives = 157/217 (71%), Gaps = 8/217 (3%)
Frame = +1
Query: 435 HTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLS 494
+T E++ SS+++FF + L + + GI P+GLF+PII+ G+ YGRL+GIL+G TN+
Sbjct: 7 NTPLEYQPSSIIIFFALYCILGLITFGIAVPSGLFLPIILMGSGYGRLLGILMGPHTNID 186
Query: 495 NGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYD 554
GL+AVLGAASL+ GSMR TVSLCVI LELTNNLLLLP+ M+VL+++K+V + FN ++Y+
Sbjct: 187 QGLFAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMIVLLIAKTVGDSFNPSIYE 366
Query: 555 LIMKAKGLPYLETHAEPYMRQLTVGDV--VTGPLQMFNGIEKVRNIVFILRTTAHNGFPV 612
+I+ KGLP+++ + EP+MR LTVG++ V + F G+EKV NIV L+ T HNGFPV
Sbjct: 367 IILHLKGLPFMDANPEPWMRNLTVGELVDVKPAVVSFKGVEKVANIVNALKNTTHNGFPV 546
Query: 613 ID------EPPGSEAPILFGIILRHHLTTLLKKKAFL 643
+D +EA L GIILR HL +LKKK FL
Sbjct: 547 MDCGLVPTTGVANEATELHGIILRAHLIQVLKKKWFL 657
>BG359405 similar to GP|4768916|gb| CLC-Nt2 protein {Nicotiana tabacum},
partial (17%)
Length = 421
Score = 124 bits (311), Expect = 2e-28
Identities = 67/138 (48%), Positives = 87/138 (62%)
Frame = +1
Query: 10 ESEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQIL 69
ES PL PLL R++ S++ +A+VG V IESLDYEI EN+ FK DWRSR VQ+L
Sbjct: 16 ESNPL-NEPLLKRNRTL--SSNPLALVGEKVSYIESLDYEINENDLFKHDWRSRSRVQVL 186
Query: 70 QYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNL 129
QYI +KWLL F++GL+ G I NLAVEN+AG K + + + R++ F F N
Sbjct: 187 QYIFLKWLLAFLVGLLTGIIATLINLAVENIAGYKLLAVLKYIHKERYLTGFLYFTGINF 366
Query: 130 SLTLFASIITAFIAPTAA 147
LT A+I+ APTAA
Sbjct: 367 RLTFVAAILCVCFAPTAA 420
>TC220055 homologue to GB|AAC26247.2|6382041|F9D12 Arabidopsis thaliana CLC-d
chloride channel protein (GB:Z71450); coded for by A.
thaliana cDNA T44764 {Arabidopsis thaliana;} , partial
(18%)
Length = 891
Score = 112 bits (281), Expect = 5e-25
Identities = 67/152 (44%), Positives = 91/152 (59%), Gaps = 5/152 (3%)
Frame = +2
Query: 599 VFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAF-----LPSPVANSYDV 653
V ILR+ HNGFPVID E P++ G++LR HL +L+ K LPS
Sbjct: 2 VSILRSNKHNGFPVIDHTRSGE-PLVIGLVLRSHLLVILQSKVDFQHSPLPSDPRGGGRS 178
Query: 654 VRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILF 713
+R S +FAK S + + I+DI L+ +++ M++DL PF N SPY V E MSL K LF
Sbjct: 179 IRH-DSGEFAKPVSSKGICIDDIHLSSDDLEMYIDLAPFLNPSPYIVPEDMSLTKVYNLF 355
Query: 714 REVGLRHLLVIPKIPGRSPVVGILTRHDFTPE 745
R++GLRHL V+P+ S VVG++TR D E
Sbjct: 356 RQLGLRHLFVVPR---PSCVVGLITRKDLLIE 442
>BE659780 similar to GP|4768916|gb| CLC-Nt2 protein {Nicotiana tabacum},
partial (20%)
Length = 644
Score = 80.1 bits (196), Expect(2) = 2e-24
Identities = 49/120 (40%), Positives = 70/120 (57%), Gaps = 7/120 (5%)
Frame = -2
Query: 603 RTTAHNGFPVID------EPPGSEAPILFGIILRHHLTTLLKKKAFLPSPV-ANSYDVVR 655
+ HNG V+D +EA L GI R +LKKK FL ++V
Sbjct: 643 KXXTHNGXXVMDCGLVPTTGVANEATELHGIXXRAXXIQVLKKKWFLKERRRTEEWEVRE 464
Query: 656 KFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFRE 715
KF+ + A++ IED+ +T+EEM MFVDLHP TN +P+TV+E+MS+AKA++LFR+
Sbjct: 463 KFTWVELAEREG----NIEDVAVTKEEMEMFVDLHPLTNTTPFTVLESMSVAKAMVLFRQ 296
Score = 51.6 bits (122), Expect(2) = 2e-24
Identities = 28/60 (46%), Positives = 39/60 (64%), Gaps = 2/60 (3%)
Frame = -1
Query: 716 VGLRHLLVIPKIP--GRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLRFWQTFLEKIL 773
+GLRH+LV+PK G SPV+GILTR D +IL + P L KS+ K ++F + +K L
Sbjct: 296 IGLRHMLVVPKYQASGVSPVIGILTRQDLLAYNILTVFPHLAKSKRK*IKFQ*IYWDKSL 117
>TC220688 similar to UP|Q9XF71 (Q9XF71) CLC-Nt2 protein, partial (13%)
Length = 496
Score = 105 bits (263), Expect = 6e-23
Identities = 56/103 (54%), Positives = 70/103 (67%)
Frame = +3
Query: 10 ESEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQIL 69
ES PL PLL R++ S++ +A+VGA V IESLDYEI EN+ FKQDWRSR Q+L
Sbjct: 195 ESNPL-NEPLLKRNRTL--SSNPLALVGAKVSYIESLDYEINENDLFKQDWRSRSRTQVL 365
Query: 70 QYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMM 112
QYI KW L F++GL+ G I NLAVEN+AG KF+ N +
Sbjct: 366 QYIFWKWTLAFLVGLLTGVIATLINLAVENIAGYKFLAVVNFI 494
>BF071148 weakly similar to GP|1742959|emb CLC-d chloride channel protein
{Arabidopsis thaliana}, partial (14%)
Length = 439
Score = 88.6 bits (218), Expect = 1e-17
Identities = 52/127 (40%), Positives = 79/127 (61%), Gaps = 1/127 (0%)
Frame = +2
Query: 489 ERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVF 548
++ N+ G YA+LGAAS LGG T+SLCVIM+EL++NL PLIM+ L++SK+ + F
Sbjct: 29 KKLNID*GTYAMLGAASFLGG*KWMTLSLCVIMVELSDNLGYSPLIMLELLISKAEGDAF 208
Query: 549 NANVYDLIMKAKGLPYLETHAEPYMRQLTVGD-VVTGPLQMFNGIEKVRNIVFILRTTAH 607
N +VYD + KG+ L++ + M+ +T + +G + F + V + ILR+ H
Sbjct: 209 NESVYDEQAQLKGIALLQSMPKYDMQNMTAHESCESGMVGSFPHVVDVSVLFNILRSNRH 388
Query: 608 NGFPVID 614
N F VID
Sbjct: 389 NDFTVID 409
>AW100482 similar to PIR|T52107|T52 anion channel protein CLC-a [validated] -
Arabidopsis thaliana, partial (16%)
Length = 408
Score = 75.9 bits (185), Expect(2) = 7e-15
Identities = 38/74 (51%), Positives = 50/74 (67%)
Frame = +3
Query: 447 VFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLSNGLYAVLGAASL 506
+ F + L + + GI P+ LF+PII G+ Y RL+GIL+G TN+ GL+AVL AASL
Sbjct: 6 ILFALHCILGLITFGIAVPSWLFLPIIFMGSGYRRLLGILMGPHTNIDQGLFAVLAAASL 185
Query: 507 LGGSMRTTVSLCVI 520
+ GSMR T SL VI
Sbjct: 186 MAGSMRMTASL*VI 227
Score = 23.5 bits (49), Expect(2) = 7e-15
Identities = 10/20 (50%), Positives = 16/20 (80%)
Frame = +1
Query: 522 LELTNNLLLLPLIMMVLVVS 541
LEL ++LLLLP+ M L+++
Sbjct: 232 LELDHDLLLLPITMHCLLIA 291
>TC229696 similar to UP|Q9XF71 (Q9XF71) CLC-Nt2 protein, partial (8%)
Length = 453
Score = 76.3 bits (186), Expect = 5e-14
Identities = 38/65 (58%), Positives = 51/65 (78%), Gaps = 2/65 (3%)
Frame = +3
Query: 696 SPYTVVETMSLAKALILFREVGLRHLLVIPK--IPGRSPVVGILTRHDFTPEHILGMHPF 753
+P+TV+E+MS+AKA+ LFR+VGLRHLLV+PK G SPV+GILTR D ++IL + P
Sbjct: 6 TPFTVLESMSVAKAMTLFRQVGLRHLLVVPKYQASGISPVIGILTRQDLLAQNILTVFPH 185
Query: 754 LVKSR 758
L KS+
Sbjct: 186 LAKSK 200
>AI938566
Length = 215
Score = 65.9 bits (159), Expect = 7e-11
Identities = 26/41 (63%), Positives = 33/41 (80%)
Frame = +2
Query: 720 HLLVIPKIPGRSPVVGILTRHDFTPEHILGMHPFLVKSRWK 760
H+ V+PK GR PVVGILTRHDF PEH+LG+HP ++ +WK
Sbjct: 8 HMCVVPKSQGRPPVVGILTRHDFMPEHVLGLHPDIMPHKWK 130
>TC223196 similar to UP|Q9XF71 (Q9XF71) CLC-Nt2 protein, partial (8%)
Length = 413
Score = 63.9 bits (154), Expect = 3e-10
Identities = 31/61 (50%), Positives = 38/61 (61%)
Frame = +3
Query: 116 RFMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVK 175
R++ F F N LT A+I+ APTAAG GI E+KAYLNGVD P +F TL VK
Sbjct: 63 RYLTGFLYFTGINFVLTFVAAILCVCFAPTAAGPGIPEIKAYLNGVDTPNMFGATTLIVK 242
Query: 176 I 176
+
Sbjct: 243 V 245
>TC228289
Length = 1798
Score = 48.1 bits (113), Expect = 1e-05
Identities = 71/313 (22%), Positives = 127/313 (39%), Gaps = 11/313 (3%)
Frame = +1
Query: 233 LIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLS 292
L+ G+AAGI++ F APV G FA+E + R FTTA I L ++I +S
Sbjct: 28 LVAAGAAAGISSGFNAPVAGCFFAIETVLRPLRAEN--SPPFTTA--MIILASVISSTVS 195
Query: 293 DKC-GLFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVI 351
+ G+ + +D SA+ ++P IL ++ G++ + +++ I
Sbjct: 196 NVLQGIQSAFTIPEYDLKSAA------ELPLYLILGMLCGVISVALTRLVAWFTKLFKTI 357
Query: 352 NEKGTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPN 411
+K FG+P + CP +G G
Sbjct: 358 QDK--------------------FGIPTVV------------CPALGGFGA----GIIAL 429
Query: 412 HYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVP 471
Y G+ F ++ +R S + L+ ++ +I L S G+V GL+ P
Sbjct: 430 KYPGILYWGFTNVEEILRTGKSASAPGIWLLAQLVAAKVIATALCKGS-GLV--GGLYAP 600
Query: 472 IIVTGASYGRLVG---------ILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIML 522
++ GA+ G + G + G YA++G A+ L + ++ +++
Sbjct: 601 SLMIGAAAGAVFGGFSAEVINSAIPGNTAVAQPPAYALVGMAATLASACSVPLTSVLLLF 780
Query: 523 ELTNNL-LLLPLI 534
ELT + +LLPL+
Sbjct: 781 ELTKDYRILLPLM 819
>TC214251
Length = 477
Score = 32.7 bits (73), Expect = 0.62
Identities = 15/27 (55%), Positives = 20/27 (73%)
Frame = +1
Query: 115 RRFMFAFFIFFASNLSLTLFASIITAF 141
+RF F FF+FF LSL+ F S+I+AF
Sbjct: 34 QRFSFYFFLFFLCTLSLS*FFSLISAF 114
>TC225943 similar to UP|NU4C_LOTJA (Q9BBP3) NAD(P)H-quinone oxidoreductase
chain 4, chloroplast (NAD(P)H dehydrogenase, chain 4)
(NADH-plastoquinone oxidoreductase chain 4) , partial
(86%)
Length = 1432
Score = 30.8 bits (68), Expect = 2.4
Identities = 21/55 (38%), Positives = 27/55 (48%)
Frame = +1
Query: 323 VFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGL 377
V IL +VGG L LF NKV++ Y TIC + LI+ + C F L
Sbjct: 52 VVILPIVGGSLIFLFPHKGNKVIKWY-------TICICLIDLLITSYVFCYHFEL 195
>TC205407 similar to UP|Q9XHD3 (Q9XHD3) PHAP2B protein, partial (46%)
Length = 954
Score = 30.4 bits (67), Expect = 3.1
Identities = 16/63 (25%), Positives = 28/63 (44%)
Frame = -2
Query: 349 NVINEKGTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQC 408
++ E I L L +I + C+L G+ W C P ++ PCP + + +
Sbjct: 755 SLFKEGSPITELILVSIIELLPGCML*GMDWN*RC---PLFSLGPCPGVAMPRLRSRS*L 585
Query: 409 PPN 411
PP+
Sbjct: 584 PPS 576
>AW831830 similar to GP|15912249|gb| AT5g54590/MRB17_9 {Arabidopsis
thaliana}, partial (36%)
Length = 331
Score = 30.4 bits (67), Expect = 3.1
Identities = 27/89 (30%), Positives = 39/89 (43%), Gaps = 7/89 (7%)
Frame = -3
Query: 131 LTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGS---- 186
L L A +TA ++P ++ A G +AP TV LCV S+ A SGS
Sbjct: 275 LELVARTLTATVSPV-------DI*AL*TGPNAPSPMTVVKLCVAFCKSLYAYSGSPDAG 117
Query: 187 --LVIGKAGPMVHTG-ACVAALLGQGGSK 212
L++ P G C++ +G G K
Sbjct: 116 IMLLLLFLNPSSQKGMPCLSDFMGDSGPK 30
>TC225683 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit 4-like protein
(26S proteasome subunit AtRPT2a), partial (97%)
Length = 1513
Score = 29.6 bits (65), Expect = 5.3
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = +1
Query: 105 FVTTSNMMLERRFMFAFFIFFASNLSLTLFASIIT 139
F+T + ER F+F F + + S LS L ++I+T
Sbjct: 1375 FITVFLLFCERMFVF*FVVVYGSTLSRDLISTIVT 1479
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.328 0.143 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,528,847
Number of Sequences: 63676
Number of extensions: 596621
Number of successful extensions: 4561
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 4467
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4546
length of query: 776
length of database: 12,639,632
effective HSP length: 105
effective length of query: 671
effective length of database: 5,953,652
effective search space: 3994900492
effective search space used: 3994900492
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144657.4


