

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.4 + phase: 0
(472 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
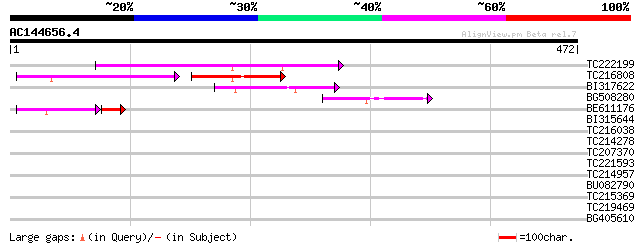
Score E
Sequences producing significant alignments: (bits) Value
TC222199 121 8e-28
TC216808 66 1e-22
BI317622 59 4e-09
BG508280 58 1e-08
BE611176 45 2e-05
BI315644 31 1.0
TC216038 similar to UP|Q9IZF4 (Q9IZF4) Aphid transmission P5, pa... 30 2.3
TC214278 similar to UP|P93704 (P93704) HMG-1, partial (98%) 30 2.3
TC207370 similar to UP|Q93ZI7 (Q93ZI7) AT5g35440/MOK9_2, partial... 30 3.0
TC221593 similar to UP|Q6NN02 (Q6NN02) At4g14450, partial (30%) 29 5.2
TC214957 similar to GB|BAA97483.1|8885553|AB025604 ripening-rela... 28 6.8
BU082790 28 6.8
TC215369 UP|O22465 (O22465) GH1 protein (Fragment), complete 28 6.8
TC219469 similar to UP|Q9ZS03 (Q9ZS03) RNA helicase (Fragment), ... 28 6.8
BG405610 28 8.9
>TC222199
Length = 691
Score = 121 bits (303), Expect = 8e-28
Identities = 75/226 (33%), Positives = 113/226 (49%), Gaps = 19/226 (8%)
Frame = +2
Query: 72 DYQLVPTLEEYSHWVGLPVLDQVPFHGLEPGPKIPAIANALHLEIADIKNKFITRAGLQC 131
D+QLVPT+EE+ +G P+ ++ P+ P + I + + TR G+
Sbjct: 5 DFQLVPTIEEFEEILGCPLGERKPYLSSGCLPSLSRIPTMVKDSARGLDRVKQTRNGIAG 184
Query: 132 LPYNFLYQKATICFERSETDAFEAILALLIYGIVLFPNVDKFVDMNAIQIFLT-----QN 186
LP +L KA + + F +LALLI+G+VLFPNVD VD+ AI FL ++
Sbjct: 185 LPRKYLEDKARGIANQGDWVPFMDVLALLIFGVVLFPNVDGLVDLAAIDAFLAYHHSKES 364
Query: 187 PVPTLLADTYVSIHERTDKERGTIVCCAPLLHIWITSHL-------------PRPKVKPE 233
PV +LAD + + + R +K I+CC P L +W+ SHL R +
Sbjct: 365 PVVAVLADLFDTFNRRYEKSSARIICCLPALCVWLVSHLFQQDTRRPCPLLSHRSCTEKR 544
Query: 234 YLPWSQKLMTLTPNDIVWFNPTCD-PELIIDSCGDFNNVPLLGTRG 278
+ W Q L + I WF + E ++ SCG + N+PL+GTRG
Sbjct: 545 RIDWDQLLAGIGGRTISWFPRWKEGKEGVLFSCGRYPNIPLVGTRG 682
>TC216808
Length = 1144
Score = 66.2 bits (160), Expect(2) = 1e-22
Identities = 36/83 (43%), Positives = 51/83 (61%), Gaps = 5/83 (6%)
Frame = +3
Query: 152 AFEAILALLIYGIVLFPNVDKFVDMNAIQIFLT-----QNPVPTLLADTYVSIHERTDKE 206
+F ILALLI+G+VLFPN++ VD+ AI FL ++PV +LA Y + +K
Sbjct: 591 SFIDILALLIFGVVLFPNMEGLVDLAAIDAFLAFHHGKESPVVAILA-MYDTFDRGCEKS 767
Query: 207 RGTIVCCAPLLHIWITSHLPRPK 229
IVCCA L++W+ SHL P+
Sbjct: 768 SARIVCCALALYVWLVSHLFSPR 836
Score = 58.5 bits (140), Expect(2) = 1e-22
Identities = 37/138 (26%), Positives = 67/138 (47%), Gaps = 2/138 (1%)
Frame = +2
Query: 6 RRTHRYEFKKPDLESLRNLGRMVKNPEH--FRNRYGYLLNILRTNVDEGLINTLVQFYDP 63
+R ++ + K ++ SL LG+++ + FR YG + ++ R V I +L Q+YD
Sbjct: 146 KRFYQVKIKSLEVTSLGELGQLMGQLQR*AFRKAYGKIWDLARVEVSMEPIASLSQYYDQ 325
Query: 64 LYHCFTFSDYQLVPTLEEYSHWVGLPVLDQVPFHGLEPGPKIPAIANALHLEIADIKNKF 123
CFTF D+QLVPT++E G + + P+ P +A + + ++
Sbjct: 326 PLRCFTFEDFQLVPTVKELEEIRGCQLGGRKPYLFSGFYPSTTRVAKVVKISAQELNRVK 505
Query: 124 ITRAGLQCLPYNFLYQKA 141
R G+ +P L ++A
Sbjct: 506 QNRNGVVGVPRKCLEERA 559
>BI317622
Length = 370
Score = 59.3 bits (142), Expect = 4e-09
Identities = 40/124 (32%), Positives = 57/124 (45%), Gaps = 20/124 (16%)
Frame = -2
Query: 171 DKFVDMNAIQIFLTQN-----PVPTLLADTYVSIHERTDKERGTIVCCAPLLHIWITSHL 225
D VD+ AI FL + P+ +LAD Y + + R +K IVCC P L++W+ SHL
Sbjct: 369 DGLVDLAAIGAFLAYHDYKEIPIVAMLADLYDTFNRRCEKSSTRIVCCTPALYVWLVSHL 190
Query: 226 PRPKVKPEYLP--------------WSQKLMTLTPNDIVWFNPTCDPEL-IIDSCGDFNN 270
R +V+ P W Q L + WF + ++ SCG F N
Sbjct: 189 FRQEVR-HACPLESHCSCTEKGGENWGQLLANKEGASVNWFPRWKEGRTRVLISCGVFLN 13
Query: 271 VPLL 274
VPL+
Sbjct: 12 VPLM 1
>BG508280
Length = 418
Score = 57.8 bits (138), Expect = 1e-08
Identities = 39/100 (39%), Positives = 54/100 (54%), Gaps = 8/100 (8%)
Frame = +1
Query: 261 IIDSCGDFNNVPLLGTRGGISYSPVLARRQFGFYM-------EMKPVYLILDRDFFLYKK 313
++ SCG F NVPL+GTRG ISY+PVLA +Q G+ M E+ PV + R F K
Sbjct: 70 VLISCGGFPNVPLMGTRGCISYNPVLAIKQLGYPM*GVPLEDELAPV---ISRGF---NK 231
Query: 314 DDANQRAQFKKAWYSIIRKDRNQLG-NRSVIAHEAYVKWV 352
+ + +KAW + +KD+ G N I Y KW+
Sbjct: 232 TNVETLQKVRKAWEVLQKKDKELRGSNNGPIG--GYRKWL 345
>BE611176
Length = 424
Score = 45.1 bits (105), Expect(2) = 2e-05
Identities = 23/72 (31%), Positives = 40/72 (54%), Gaps = 2/72 (2%)
Frame = -2
Query: 6 RRTHRYEFKKPDLESLRNLGRMVK--NPEHFRNRYGYLLNILRTNVDEGLINTLVQFYDP 63
+R ++ + K D+ S++ LGR++ + FR Y +L++ + I +L Q+YD
Sbjct: 360 KRFYQVKIKGLDITSIKELGRLMGPLQMQAFRKAYEKILDLTIAEISTEAIASLTQYYD* 181
Query: 64 LYHCFTFSDYQL 75
CFTF D+QL
Sbjct: 180 PLRCFTFGDFQL 145
Score = 21.2 bits (43), Expect(2) = 2e-05
Identities = 7/20 (35%), Positives = 13/20 (65%)
Frame = -1
Query: 77 PTLEEYSHWVGLPVLDQVPF 96
PT+EE+ +G P+ + P+
Sbjct: 142 PTVEEFEEILGCPLRGRKPY 83
>BI315644
Length = 421
Score = 31.2 bits (69), Expect = 1.0
Identities = 11/22 (50%), Positives = 15/22 (68%)
Frame = +1
Query: 204 DKERGTIVCCAPLLHIWITSHL 225
+K IVCCA L++W+ SHL
Sbjct: 1 EKSSARIVCCALALYVWLVSHL 66
Score = 29.6 bits (65), Expect = 3.0
Identities = 14/24 (58%), Positives = 17/24 (70%)
Frame = +3
Query: 276 TRGGISYSPVLARRQFGFYMEMKP 299
TRG I+Y+PVLA RQ G+ M P
Sbjct: 276 TRGCINYNPVLAIRQLGYPMRGVP 347
>TC216038 similar to UP|Q9IZF4 (Q9IZF4) Aphid transmission P5, partial (3%)
Length = 1754
Score = 30.0 bits (66), Expect = 2.3
Identities = 23/82 (28%), Positives = 36/82 (43%), Gaps = 9/82 (10%)
Frame = +3
Query: 386 EGYQKQLDIERRENSMWEVKYRKKKQEYDTV-KNLLDQQIQANCKEKNENARLKASIQRK 444
+G + D E++EN E K KKK++ D K + + Q Q N + N L +
Sbjct: 1059 KGVETDGDGEKKENVPAESKTSKKKKKKDKASKEVKESQDQPNSSDTNNGPDLATGAENV 1238
Query: 445 EDF--------LDKICPGRKKR 458
ED L K+ +KK+
Sbjct: 1239 EDTSTVDVKERLKKVASTKKKK 1304
>TC214278 similar to UP|P93704 (P93704) HMG-1, partial (98%)
Length = 932
Score = 30.0 bits (66), Expect = 2.3
Identities = 14/60 (23%), Positives = 31/60 (51%)
Frame = +1
Query: 387 GYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKASIQRKED 446
G +K + E + + KKKQEY+ + ++Q++ E++E+ + K+ + E+
Sbjct: 502 GGEKWKSLSDAEKAPFVATAEKKKQEYEKTISAYNKQLEGKNSEEDESDKSKSEVNDDEE 681
>TC207370 similar to UP|Q93ZI7 (Q93ZI7) AT5g35440/MOK9_2, partial (15%)
Length = 1647
Score = 29.6 bits (65), Expect = 3.0
Identities = 15/47 (31%), Positives = 27/47 (56%)
Frame = +3
Query: 312 KKDDANQRAQFKKAWYSIIRKDRNQLGNRSVIAHEAYVKWVIDRANK 358
K ++ + Q ++Y +RK NQL ++V+A+ AYV+ +D K
Sbjct: 672 KGGNSQELVQNSLSYYENVRKRENQLVKQAVLANLAYVELELDNPVK 812
>TC221593 similar to UP|Q6NN02 (Q6NN02) At4g14450, partial (30%)
Length = 514
Score = 28.9 bits (63), Expect = 5.2
Identities = 17/51 (33%), Positives = 26/51 (50%)
Frame = +1
Query: 353 IDRANKWKMPYPRQRLVTSTVSAIPLPLPPESLEGYQKQLDIERRENSMWE 403
I+RA +W + P + S S PLPLPP+ + Q+Q E+ W+
Sbjct: 193 INRALEWNVAIPLLSPLAS--SPPPLPLPPQKEDPPQQQRPTEKVVFKKWQ 339
>TC214957 similar to GB|BAA97483.1|8885553|AB025604 ripening-related
protein-like; hydrolase-like {Arabidopsis thaliana;} ,
partial (77%)
Length = 1262
Score = 28.5 bits (62), Expect = 6.8
Identities = 33/126 (26%), Positives = 51/126 (40%), Gaps = 12/126 (9%)
Frame = +3
Query: 59 QFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPVLDQVPFH-----GLEPGPKIPAIANALH 113
Q P Y C F L TL + L + + G++P KI ++N L+
Sbjct: 135 QVQRPKYDCLLFD---LDDTLYPLKSGLAKSCLQNIKHYMVEKLGIDPS-KIDDLSNLLY 302
Query: 114 LEIADIKNKFITRAGLQCLPYNFLYQK------ATICFERSETD-AFEAILALLIYGIVL 166
KN T AGL+ + Y+F Y + + +E + D +L L Y ++
Sbjct: 303 ------KNYGTTMAGLRAIGYDFDYDEYHSFVHGRLPYENLKPDPVLRNLLLSLPYRKLI 464
Query: 167 FPNVDK 172
F N DK
Sbjct: 465 FTNADK 482
>BU082790
Length = 431
Score = 28.5 bits (62), Expect = 6.8
Identities = 15/46 (32%), Positives = 22/46 (47%), Gaps = 4/46 (8%)
Frame = -3
Query: 213 CAPLLHIWITSHLPRPKVK----PEYLPWSQKLMTLTPNDIVWFNP 254
C P+ WI + P PK+K P+ PW TL+ + F+P
Sbjct: 126 CPPVPQFWIGAPFPSPKMKKLTFPKQFPW-----TLSCPQVFLFSP 4
>TC215369 UP|O22465 (O22465) GH1 protein (Fragment), complete
Length = 1555
Score = 28.5 bits (62), Expect = 6.8
Identities = 15/41 (36%), Positives = 23/41 (55%), Gaps = 1/41 (2%)
Frame = -1
Query: 370 TSTVSAIPLPLPPESLEGYQKQLDIERREN-SMWEVKYRKK 409
T+ SA P PLPP + Y K + +EN + E++ R+K
Sbjct: 220 TTRCSAPPPPLPPPAATSYSKHTNKPNKENLKL*EIRDRRK 98
>TC219469 similar to UP|Q9ZS03 (Q9ZS03) RNA helicase (Fragment), partial
(66%)
Length = 1062
Score = 28.5 bits (62), Expect = 6.8
Identities = 47/192 (24%), Positives = 83/192 (42%), Gaps = 9/192 (4%)
Frame = +3
Query: 249 IVWFNPTCDPELIIDSCGDFNNVPLLGTRGGISYSPVLARRQFGFYMEMKPVYLILDRDF 308
IV ++P DP + I G LG +G ++ V + Y+E + R
Sbjct: 336 IVQYDPPQDPNVFIHRVG---RTARLGKQG---HAVVFLLPKEESYVEFLRI-----RRV 482
Query: 309 FLYKKDDANQRAQFKKAWYSIIRKDRNQL--GNRSVIAH-EAYVKWVIDRANKWKMPYPR 365
L ++ A++ + S +KDR+ + G ++ +++ AY + +WK
Sbjct: 483 PLQERICADEASDVVPQIRSAAKKDRDVMEKGIKAFVSYIRAYKEHHCSYIFRWK-ELEI 659
Query: 366 QRLVTSTVSAIPLPLPPE------SLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNL 419
+L T + LP PE S++G++ DI ++ ++KYR K +E KNL
Sbjct: 660 GKLATG-FGLLQLPSMPEVKHHSLSIDGFEPVEDI-----NLGDIKYRDKSREKQRKKNL 821
Query: 420 LDQQIQANCKEK 431
Q + KEK
Sbjct: 822 ---QAKKEAKEK 848
>BG405610
Length = 406
Score = 28.1 bits (61), Expect = 8.9
Identities = 9/31 (29%), Positives = 16/31 (51%)
Frame = +3
Query: 323 KKAWYSIIRKDRNQLGNRSVIAHEAYVKWVI 353
K WY + + DRN+ + + +H WV+
Sbjct: 126 KGCWYHVSKMDRNKSRVKDISSHNCIASWVL 218
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.140 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,711,451
Number of Sequences: 63676
Number of extensions: 353899
Number of successful extensions: 1862
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 1847
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1857
length of query: 472
length of database: 12,639,632
effective HSP length: 101
effective length of query: 371
effective length of database: 6,208,356
effective search space: 2303300076
effective search space used: 2303300076
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144656.4


