

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.13 + phase: 0 /pseudo/partial
(1235 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
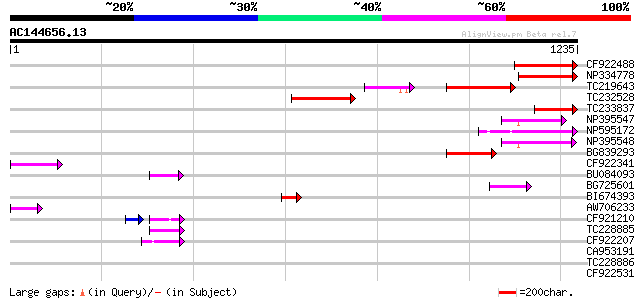
Score E
Sequences producing significant alignments: (bits) Value
CF922488 196 4e-50
NP334778 reverse transcriptase [Glycine max] 186 5e-47
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 168 2e-41
TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotei... 152 1e-36
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 132 1e-30
NP395547 reverse transcriptase [Glycine max] 130 5e-30
NP595172 polyprotein [Glycine max] 118 2e-26
NP395548 reverse transcriptase [Glycine max] 117 2e-26
BG839293 108 1e-23
CF922341 96 1e-19
BU084093 60 6e-09
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 55 1e-07
BI674393 54 4e-07
AW706233 54 6e-07
CF921210 47 2e-05
TC228885 47 7e-05
CF922207 44 3e-04
CA953191 42 0.002
TC228886 39 0.014
CF922531 38 0.024
>CF922488
Length = 741
Score = 196 bits (499), Expect = 4e-50
Identities = 95/136 (69%), Positives = 108/136 (78%)
Frame = +3
Query: 1100 VPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSP 1159
V K+DGKV MCVD+RDLN ASPKD FPL HI+VLVDN F FMDGFSGYNQIK++P
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 1160 EDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKD 1219
ED EKT+FIT WGTFCYK M GL N GATYQR M LF DM+HKE+EVY+DDMIVKS+
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 1220 EEQHVEYLTKMFERLR 1235
EE+H+ L K+F RLR
Sbjct: 363 EEEHLVNLRKLFRRLR 410
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 186 bits (472), Expect = 5e-47
Identities = 87/128 (67%), Positives = 102/128 (78%)
Frame = +3
Query: 1108 RMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSF 1167
RMCVD+RDLN+ASPKDNFPL HID+L+ N A +F FMDGFSGYNQIKM+PED EKT+F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 1168 ITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYL 1227
IT WGTFCYKVM GL N GATY R M LF DM+HKE+E YVD+MI KS+ EE+H+ L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 1228 TKMFERLR 1235
+F +LR
Sbjct: 363 QNLFGQLR 386
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein (Fragment),
partial (8%)
Length = 1320
Score = 168 bits (425), Expect = 2e-41
Identities = 77/152 (50%), Positives = 108/152 (70%), Gaps = 2/152 (1%)
Frame = +3
Query: 952 NFEFPMYEAEAEEGDDI--PYEITRLLEQEKKAIQPHQEEIEFINIGTEENKREIKVGAA 1009
+FE M + E E +D+ P E+ R++ E + + PHQEE E +++G+ KRE+K+G
Sbjct: 864 DFEQKMNQTEDEGNEDVGLPPELERMVAHEDQEMGPHQEETELVDLGSGSGKREVKIGTG 1043
Query: 1010 LEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMA 1069
+ +++++I LLR+Y DIFAWSY+DMPGL IV+HR+P PEC PV+QKLRR P+ +
Sbjct: 1044 ITAPIREELIILLRDYQDIFAWSYQDMPGLSSDIVQHRLPLNPECSPVKQKLRRMKPETS 1223
Query: 1070 LKIKNEVQKQIDAGFLMTVEYPEWVANIVPVP 1101
LKIK EV+K DAGFL YP+WVANIVP+P
Sbjct: 1224 LKIKEEVKK*FDAGFLAVARYPKWVANIVPIP 1319
Score = 65.5 bits (158), Expect = 1e-10
Identities = 47/125 (37%), Positives = 56/125 (44%), Gaps = 15/125 (12%)
Frame = +1
Query: 773 PWIHDAGAVTSTLHQNFKFVKNGKLVTVHGEEAYLVSQLSSFSCIEAGSAE-GTAFQGLT 831
PWIH G V STLHQ KFV G LV V GEE LVS SS +EA TAFQ
Sbjct: 1 PWIHSVGVVPSTLHQKLKFVVEGHLVIVSGEEDILVSCPSSMPYVEAAEESLETAFQSFE 180
Query: 832 IEGMELKKTGTAMASLKD-----AQKAVQEGQAAGRG---------KLIQLRENKHKEGL 877
+ + + L D A+ + G G G LI + N+ K GL
Sbjct: 181 VVSISSVDSLFGQPCLSDAAVMMARVMLGNGYEPGMGLGKDNGGITSLINTQGNRGKYGL 360
Query: 878 GFSPT 882
G+ PT
Sbjct: 361 GYKPT 375
>TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotein
(Fragment), partial (3%)
Length = 449
Score = 152 bits (383), Expect = 1e-36
Identities = 72/139 (51%), Positives = 105/139 (74%)
Frame = +1
Query: 614 SKISVLSLLLSSEAHRNTLLKVLEQAYVDHEVTMDRFGSIVGNITACNNLWFSEDELPEA 673
+++S+L LL+SSE HR L+KVL +A+V +++++ FG +V NITA N L F+E+E+P
Sbjct: 31 ARVSLLELLMSSEPHRALLVKVLNEAHVAQDISVEGFGGLVNNITANNYLAFAEEEIPAE 210
Query: 674 GKYHNLALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFLVKAFDG 733
G+ HN ALH+SV C +++ VL+D G SLNVMPKSTLD+L + + L+ S+ +V+AFDG
Sbjct: 211 GRGHNKALHVSVKCMDHIVAKVLIDNGYSLNVMPKSTLDKLPFNASHLKPSSMVVRAFDG 390
Query: 734 SRKNVLGEIDLPMTIGPET 752
+R+ V GEIDLP+ IGP T
Sbjct: 391 TRREVRGEIDLPVQIGPHT 447
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 132 bits (331), Expect = 1e-30
Identities = 62/93 (66%), Positives = 73/93 (77%)
Frame = +2
Query: 1143 FFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMI 1202
F FMDGFSGYNQI M+ ED EKT+F+T WGTF Y+VM GL N GATYQR M LFHDM+
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 1203 HKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
HKE+EVYVDDMI KS+ E +H+ L K+F RL+
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQ 280
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 130 bits (326), Expect = 5e-30
Identities = 62/159 (38%), Positives = 95/159 (58%), Gaps = 18/159 (11%)
Frame = +1
Query: 1072 IKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV------------------RMCVDF 1113
++ EV K ++AG + + WV+ + VPKK G RMC+D+
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 1114 RDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGT 1173
R LN+A+ KD++PL +D ++ A+ + F+DG+SGYNQI + P+D+EKT+F P+
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 1174 FCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDD 1212
F Y+ MP GL NA T+QR M +F DM+ K +EV++DD
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDD 477
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 118 bits (295), Expect = 2e-26
Identities = 74/215 (34%), Positives = 115/215 (53%)
Frame = +1
Query: 1021 LLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQI 1080
LL Y +FA P + +H IP K PV+ + R +I+ +Q+ +
Sbjct: 1693 LLHTYAQVFAVPASLPPQREQ---DHAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEML 1863
Query: 1081 DAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQS 1140
G + P + I+ V KKDG R C D+R LN + KD+FP+ +D L+D +
Sbjct: 1864 VQGIIQPSNSP-FSLPILLVKKKDGSWRFCTDYRALNAITVKDSFPMPTVDELLDELHGA 2040
Query: 1141 KVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHD 1200
+ F +D SGY+QI + PEDREKT+F T G + + VMP GL NA AT+Q M +F
Sbjct: 2041 QYFSKLDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQF 2220
Query: 1201 MIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
+ K V V+ DD+++ S + H+++L + + L+
Sbjct: 2221 ALRKFVLVFFDDILIYSASWKDHLKHLESVLQTLK 2325
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 117 bits (294), Expect = 2e-26
Identities = 58/180 (32%), Positives = 100/180 (55%), Gaps = 18/180 (10%)
Frame = +1
Query: 1072 IKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV------------------RMCVDF 1113
++ EV K ++ G + + WV+ ++ V KK+G ++C+D+
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 1114 RDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGT 1173
R LN+A+ KD+FPL +D +++ A + F+D + GYNQI + P+D+EK +F P+G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 1174 FCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFER 1233
F Y+ +P GL NA T+Q M +F D++ K +EV++DD V E ++ L + +R
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
>BG839293
Length = 781
Score = 108 bits (270), Expect = 1e-23
Identities = 48/110 (43%), Positives = 74/110 (66%), Gaps = 2/110 (1%)
Frame = +1
Query: 952 NFEFPMYEAEAEEGDDI--PYEITRLLEQEKKAIQPHQEEIEFINIGTEENKREIKVGAA 1009
NFE + E E +D+ P E+ R++ E + + PHQEE E +++G KRE+K+G
Sbjct: 400 NFEQETSQTEDEGNEDVGLPPELERMVAHEDQEMGPHQEETELVDLGIGSGKREVKIGTG 579
Query: 1010 LEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQ 1059
+ +++++I LL++Y DIFAWSY+DMPGL IV+H++P PEC PV+Q
Sbjct: 580 ITAPIREELIILLKDYQDIFAWSYQDMPGLSSDIVQHQLPLNPECSPVKQ 729
>CF922341
Length = 675
Score = 95.5 bits (236), Expect = 1e-19
Identities = 49/119 (41%), Positives = 68/119 (56%), Gaps = 4/119 (3%)
Frame = +1
Query: 1 MEELAK--ELRREIKANRGNGDSV--KTHDLCLVPKVDVPKKFKVPEFDRYNGLTCPHNH 56
M E+ K L ++A G D +L LVP + P KFKV +FD+Y G TCP NH
Sbjct: 289 MAEMGKLDHLEEGLRAIEGGEDYAFANLEELFLVPNIITPPKFKVLDFDKYKGTTCPKNH 468
Query: 57 IIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFN 115
+ Y +KMG Y+ ++ L+IH FQ+SL A WYT+L VH++ +L AF Y +N
Sbjct: 469 LKMYCQKMGAYAKDEELLIHSFQESLTGVAVTWYTNLEPSRVHSWKDLMVAFVRQYQYN 645
>BU084093
Length = 421
Score = 60.1 bits (144), Expect = 6e-09
Identities = 29/75 (38%), Positives = 40/75 (52%)
Frame = -1
Query: 304 FPPIPMLYAELLPTLLLRGHCTTRQDKPPPDPLPPRFRSDLKCDFHQGALGHDVKGCYAL 363
F P+PM Y +LLP+L+ + P P + + C +H G GH V+ C AL
Sbjct: 226 FTPLPMTYEDLLPSLIANHLAVVTPGRVLQPPFPKWYDPNATCKYHGGVPGHSVEKCLAL 47
Query: 364 KYIVKKLIDQGKLTF 378
KY V+ L+D G LTF
Sbjct: 46 KYKVQHLMDAGWLTF 2
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 55.5 bits (132), Expect = 1e-07
Identities = 30/90 (33%), Positives = 48/90 (53%)
Frame = -3
Query: 1046 HRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDG 1105
H++ + V Q+ R+ + + EV K A F+ + Y + ++V V K +G
Sbjct: 277 HKLAICNDVKLVTQRKRKIREERCQTV*QEVVKLAIASFIRDINYST*LFSVVMVKKPNG 98
Query: 1106 KVRMCVDFRDLNKASPKDNFPLLHIDVLVD 1135
K R+C D+ DLN A PKD +PL +ID + D
Sbjct: 97 KWRICTDYIDLN*ACPKDAYPLPNIDHMTD 8
>BI674393
Length = 152
Score = 53.9 bits (128), Expect = 4e-07
Identities = 23/44 (52%), Positives = 38/44 (86%)
Frame = +1
Query: 593 EILRLIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVL 636
E LR+I++S++K+++QL +TP+++S+L LL+SSE HR L+KVL
Sbjct: 16 EFLRIIQQSEFKVIEQLNKTPARVSLLELLMSSEPHRALLVKVL 147
>AW706233
Length = 376
Score = 53.5 bits (127), Expect = 6e-07
Identities = 28/71 (39%), Positives = 37/71 (51%), Gaps = 2/71 (2%)
Frame = -3
Query: 3 ELAKELRREIKANRGNGDSV--KTHDLCLVPKVDVPKKFKVPEFDRYNGLTCPHNHIIKY 60
E L+ KA G D +L LV + P KFKV FD+Y G TCP NH+ Y
Sbjct: 347 EKLDHLKERFKAIEGGQDYAFANLEELFLVXNIISPPKFKVLNFDKYKGTTCPKNHLKMY 168
Query: 61 VRKMGNYSDND 71
+KMG Y+ ++
Sbjct: 167 CQKMGAYAKDE 135
>CF921210
Length = 790
Score = 46.6 bits (109), Expect(2) = 2e-05
Identities = 32/78 (41%), Positives = 39/78 (49%)
Frame = +2
Query: 304 FPPIPMLYAELLPTLLLRGHCTTRQDKPPPDPLPPRFRSDLKCDFHQGALGHDVKGCYAL 363
F PIP+ YA+LL LL K PL + S+ C GAL H ++ C AL
Sbjct: 341 FTPIPVSYADLLSYLLDNSMVAITLAKVHQPPLF*GYDSNATCG---GALRHSIEHCRAL 511
Query: 364 KYIVKKLIDQGKLTFENN 381
K V+ LID G L FE N
Sbjct: 512 KRKVQGLIDAGWLKFEEN 565
Score = 20.8 bits (42), Expect(2) = 2e-05
Identities = 11/40 (27%), Positives = 12/40 (29%)
Frame = +1
Query: 252 PPSYPYAPYSQHPFFPYPLPSGQPQVPVNAVVQQMQQQPP 291
P YP Y Q PF P N Q+ P
Sbjct: 187 PSPYPPPRYPQRPFLNLPQSLSTALPMTNTTFSTNQKHQP 306
>TC228885
Length = 901
Score = 46.6 bits (109), Expect = 7e-05
Identities = 30/78 (38%), Positives = 37/78 (46%)
Frame = -3
Query: 304 FPPIPMLYAELLPTLLLRGHCTTRQDKPPPDPLPPRFRSDLKCDFHQGALGHDVKGCYAL 363
F PIP+ YA+LLP LL K P + S+ C H A G ++ AL
Sbjct: 617 FTPIPVSYADLLPYLLDNSMVAITLAKVHQPPFLREYDSNAMCACHGEAPGRSIEHYRAL 438
Query: 364 KYIVKKLIDQGKLTFENN 381
K V+ LID G L FE N
Sbjct: 437 KRKVQGLIDAGWLKFEEN 384
>CF922207
Length = 616
Score = 44.3 bits (103), Expect = 3e-04
Identities = 32/94 (34%), Positives = 42/94 (44%)
Frame = -1
Query: 288 QQPPVQQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRQDKPPPDPLPPRFRSDLKCD 347
QQ + QQ+ + P F ML L+P L K P P + S+ C
Sbjct: 601 QQKGILQQKACRVHPKFR-CHML--NLIPYPLDNSMVAITPTKVPQPPFFREYDSNATCA 431
Query: 348 FHQGALGHDVKGCYALKYIVKKLIDQGKLTFENN 381
+H GA GH ++ C K+ V LID G L FE N
Sbjct: 430 YHGGAPGHSIEHCMTPKHKV*SLIDTG*LKFEEN 329
>CA953191
Length = 422
Score = 41.6 bits (96), Expect = 0.002
Identities = 27/58 (46%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Frame = -3
Query: 762 INASYSCLLGRPW-IHDAGAVTSTLHQNFKFVKNGKLVTVHGEEAYLVSQLSSFSCIE 818
I +Y+ L GRPW IH V STLH K V +GKLV + +E LV + SS IE
Sbjct: 420 ITPTYNGLQGRPWRIHCVKLVPSTLH*K*KIVIDGKLVIIFVKEDLLVGEPSSTPYIE 247
>TC228886
Length = 748
Score = 38.9 bits (89), Expect = 0.014
Identities = 18/42 (42%), Positives = 24/42 (56%)
Frame = +3
Query: 340 FRSDLKCDFHQGALGHDVKGCYALKYIVKKLIDQGKLTFENN 381
+ S+ C +H GA GH ++ C K+ V LID G L FE N
Sbjct: 153 YDSNATCAYHGGASGHSIEHCMTPKHKV*SLIDTGWLKFEEN 278
>CF922531
Length = 602
Score = 38.1 bits (87), Expect = 0.024
Identities = 28/95 (29%), Positives = 52/95 (54%), Gaps = 2/95 (2%)
Frame = -2
Query: 1012 EWVKKKIIQLLREYPDIFAWSYEDMP-GLDP-KIVEHRIPTKPECPPVRQKLRRTHPDMA 1069
E + K+ +LL E+ DIF +++P GL P + +EH+I P + RT+P
Sbjct: 274 ETLPPKVQELLHEFGDIFP---KEIPLGLPPLRGIEHQIDLVPRASLPNRPTYRTNPQET 104
Query: 1070 LKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKD 1104
+I+++V++ ++ G++ V ++ VPKKD
Sbjct: 103 KEIESQVKELLEKGWVQE-SLSLCVVLVLLVPKKD 2
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,933,100
Number of Sequences: 63676
Number of extensions: 818482
Number of successful extensions: 5648
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 5088
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5500
length of query: 1235
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1127
effective length of database: 5,762,624
effective search space: 6494477248
effective search space used: 6494477248
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC144656.13


