

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.10 - phase: 0
(324 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
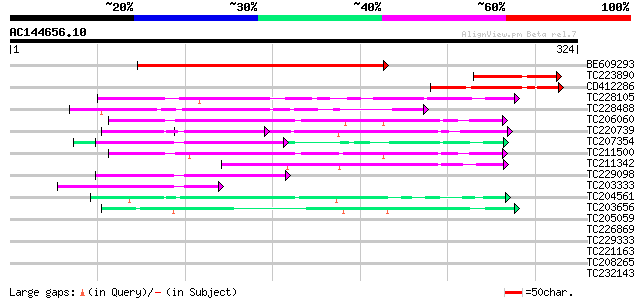
Score E
Sequences producing significant alignments: (bits) Value
BE609293 213 7e-56
TC223890 75 4e-14
CD412286 73 2e-13
TC228105 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {A... 53 2e-07
TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%) 53 2e-07
TC206060 weakly similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-... 50 1e-06
TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nu... 50 1e-06
TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat prot... 48 7e-06
TC211500 similar to UP|Q9FND4 (Q9FND4) WD-repeat protein-like, p... 47 9e-06
TC211342 similar to UP|Q9LVF2 (Q9LVF2) Arabidopsis thaliana geno... 44 8e-05
TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory pr... 44 1e-04
TC203333 similar to UP|Q8H6S5 (Q8H6S5) CTV.2, partial (54%) 41 8e-04
TC204561 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-... 41 8e-04
TC203656 similar to UP|Q9SYX2 (Q9SYX2) Phytochrome A supressor s... 41 8e-04
TC205059 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-... 40 0.002
TC226869 similar to UP|Q8H6S5 (Q8H6S5) CTV.2, partial (40%) 40 0.002
TC229333 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial ... 39 0.004
TC221163 39 0.004
TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%) 39 0.004
TC232143 similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-like, p... 38 0.005
>BE609293
Length = 436
Score = 213 bits (543), Expect = 7e-56
Identities = 105/144 (72%), Positives = 121/144 (83%), Gaps = 1/144 (0%)
Frame = +3
Query: 74 GSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKG-VND 132
GSL HS+TVTALSFY+PP+LPFPRNLVSADA G+L+IF ADGFVHL TLSVH+ +ND
Sbjct: 3 GSLHTHSATVTALSFYAPPHLPFPRNLVSADASGNLSIFHADGFVHLTTLSVHRNSAIND 182
Query: 133 LAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVN 192
LA+HPSG+ ALTV RD C A+VNLVRGRR+ C RL KEAS+V+FD SGD FFMAV E V+
Sbjct: 183 LALHPSGERALTVGRDECLAVVNLVRGRRNCCLRLGKEASLVKFDASGDQFFMAVKEKVS 362
Query: 193 VHQAEDARLLMELQCPKRVLCAAP 216
VHQ EDAR+L EL+CPK VLCA P
Sbjct: 363 VHQTEDARILFELECPKPVLCATP 434
>TC223890
Length = 440
Score = 75.1 bits (183), Expect = 4e-14
Identities = 39/50 (78%), Positives = 41/50 (82%)
Frame = +2
Query: 266 DDEPYLVASASSDGTIRAWDVRMAATEKSEPLAECKTQSRLTCLAGSSLK 315
DD PYLVASASSDG IR WDVRMAA EK PL E T++RLTCLAGSSLK
Sbjct: 2 DDNPYLVASASSDGIIRVWDVRMAAREK--PLTEHNTRARLTCLAGSSLK 145
>CD412286
Length = 292
Score = 73.2 bits (178), Expect = 2e-13
Identities = 44/76 (57%), Positives = 51/76 (66%)
Frame = -1
Query: 241 VAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDVRMAATEKSEPLAEC 300
+ YC+EE H+ RVKGIVV +D + DEPYL+A SSDG IR DVRMAA EK PL EC
Sbjct: 262 LTYCLEEGHSTRVKGIVVTTD--SDGDEPYLMAFGSSDGIIRF*DVRMAAREK--PLVEC 95
Query: 301 KTQSRLTCLAGSSLKC 316
Q + LAGS KC
Sbjct: 94 -IQF*IDVLAGSYHKC 50
>TC228105 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Arabidopsis
thaliana;} , complete
Length = 1113
Score = 52.8 bits (125), Expect = 2e-07
Identities = 62/245 (25%), Positives = 104/245 (42%), Gaps = 4/245 (1%)
Frame = +3
Query: 51 SVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADG--- 107
SV AS D + ++D+ +++++ +L S V + F P+ + A A G
Sbjct: 213 SVAASSSLDSFVRVFDVDSNATIATLEAPPSEVWQMRFD-------PKGAILAVAGGGSA 371
Query: 108 SLAIFDADGFVHLKTLSVHK-KGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCR 166
S+ ++D + + TLS+ + +G S K L+V+ G+R C
Sbjct: 372 SVKLWDTSSWELVATLSIPRPEGQKPTDKSGSKKFVLSVAWSP--------DGKRLACGS 527
Query: 167 LDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLLYTGG 226
+D S+ FDV F +H E + P R L +P LL+T
Sbjct: 528 MDGTISV--FDVPRAKF---------LHHLEGHFM------PVRSLVYSPYDPRLLFTAS 656
Query: 227 EDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDV 286
+D N+ +D + GK HA+ V + V D A +A+ SSD ++R WD+
Sbjct: 657 DDGNVHMYDAE-GKALIGTMSGHASWVLCVDVSPDGAA-------IATGSSDRSVRLWDL 812
Query: 287 RMAAT 291
M A+
Sbjct: 813 NMRAS 827
>TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%)
Length = 1303
Score = 52.8 bits (125), Expect = 2e-07
Identities = 52/208 (25%), Positives = 92/208 (44%), Gaps = 3/208 (1%)
Frame = +3
Query: 35 SYPSHLSLIKTVAVSNS--VVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPP 92
SY HLS + +A+ + V+ +GG D ++D+ + + +L+ H +TV S ++ P
Sbjct: 747 SYHGHLSGVYCLALHPTIDVLLTGGRDSVCRVWDIRSKMQIHALSGHDNTVC--SVFTRP 920
Query: 93 NLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFA 152
P +V+ D ++ ++D + TL+ HKK V +A HP + + S D+
Sbjct: 921 TDP---QVVTGSHDTTIKMWDLRYGKTMSTLTNHKKSVRAMAQHPKEQAFASASADN-IK 1088
Query: 153 MVNLVRGRRSFCCR-LDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRV 211
NL +G FC L ++ +I+ MAV+E
Sbjct: 1089KFNLPKGE--FCHNMLSQQKTIINA--------MAVNE---------------------- 1172
Query: 212 LCAAPAKNGLLYTGGEDRNITAWDLKSG 239
G++ TGG++ ++ WD KSG
Sbjct: 1173-------EGVMVTGGDNGSMWFWDWKSG 1235
Score = 35.8 bits (81), Expect = 0.027
Identities = 31/111 (27%), Positives = 49/111 (43%)
Frame = +3
Query: 39 HLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPR 98
HL K V+ V+ TI ++DL++ +LT H V L+ + F
Sbjct: 513 HLL*WKECQVNGHVLYGMHHGRTIKIWDLASGVLKLTLTGHIEQVRGLAVSNRHTYMF-- 686
Query: 99 NLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDS 149
SA D + +D + +++ H GV LA+HP+ + LT RDS
Sbjct: 687 ---SAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLALHPTIDVLLTGGRDS 830
Score = 34.3 bits (77), Expect = 0.079
Identities = 17/74 (22%), Positives = 35/74 (46%)
Frame = +3
Query: 214 AAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVA 273
A ++ +++ G+D+ + WDL+ KV ++ + G+ L+ T D ++
Sbjct: 657 AVSNRHTYMFSAGDDKQVKCWDLEQNKVI----RSYHGHLSGVYCLALHPTID----VLL 812
Query: 274 SASSDGTIRAWDVR 287
+ D R WD+R
Sbjct: 813 TGGRDSVCRVWDIR 854
>TC206060 weakly similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-like,
partial (62%)
Length = 1510
Score = 50.4 bits (119), Expect = 1e-06
Identities = 50/237 (21%), Positives = 103/237 (43%), Gaps = 9/237 (3%)
Frame = +3
Query: 57 GSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADG 116
G ++ I +D+ST L + + + S++ P + ++ +D S+ +++ DG
Sbjct: 615 GVEEAIRRWDVSTGKCLQIYEKAGAGLVSCSWF-----PCGKYILCGLSDKSICMWELDG 779
Query: 117 FVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRF 176
++DL I G+ L++ + + + N F +++ +I F
Sbjct: 780 KEVESWKGQKTLKISDLEITDDGEEILSICKANVVLLFNRETKDERF---IEEYETITSF 950
Query: 177 DVSGDSFFMAVDEI---VNVHQAE-DARLLMELQCPKRVL-----CAAPAKNGLLYTGGE 227
+S D+ F+ V+ + +++ E D +L+ + + KR C K + +G E
Sbjct: 951 SLSNDNKFLLVNLLNQEIHLWNIEGDPKLVGKYKGHKRARFIIRSCFGGLKQAFIASGSE 1130
Query: 228 DRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAW 284
D + W SG++ + H+ V + + P+++ASAS D TIR W
Sbjct: 1131DSQVYIWHRSSGELIEALT-GHSGSVNCV------SWNPANPHMLASASDDRTIRVW 1280
>TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nuclear
ribonucleoprotein Prp4 (U4/U6 snRNP 60 kDa protein) (WD
splicing factor Prp4) (hPrp4), partial (34%)
Length = 1206
Score = 50.4 bits (119), Expect = 1e-06
Identities = 54/198 (27%), Positives = 81/198 (40%), Gaps = 5/198 (2%)
Frame = +1
Query: 95 PFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMV 154
P +L +A AD + ++ LKT H + +A HPSGK T S D + +
Sbjct: 229 PVHDHLATASADRTAKYWNQGSL--LKTFEGHLDRLARIAFHPSGKYLGTASFDKTWRLW 402
Query: 155 NLVRGRRSFCCRLDKEASIVRFDVSGDSFFMA---VDEIVNVHQAEDARLLMELQCP-KR 210
++ G + S+ D A +D + V R ++ L+ K
Sbjct: 403 DIETGDE-LLLQEGHSRSVYGLAFHNDGSLAASCGLDSLARVWDLRTGRSILALEGHVKP 579
Query: 211 VLCAAPAKNGL-LYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEP 269
VL + + NG L TGGED WDL+ K Y I AH+ ++S E
Sbjct: 580 VLGISFSPNGYHLATGGEDNTCRIWDLRKKKSFYTI-PAHSN------LISQVKFEPHEG 738
Query: 270 YLVASASSDGTIRAWDVR 287
Y + +AS D T + W R
Sbjct: 739 YFLVTASYDMTAKVWSGR 792
Score = 43.9 bits (102), Expect = 1e-04
Identities = 29/96 (30%), Positives = 49/96 (50%)
Frame = +1
Query: 53 VASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIF 112
+A+GG D+T ++DL S ++ HS+ ++ + F P F LV+A D + ++
Sbjct: 616 LATGGEDNTCRIWDLRKKKSFYTIPAHSNLISQVKF-EPHEGYF---LVTASYDMTAKVW 783
Query: 113 DADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRD 148
F +KTLS H+ V + + G +TVS D
Sbjct: 784 SGRDFKPVKTLSGHEAKVTSVDVLGDGGSIVTVSHD 891
>TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat protein 5,
partial (57%)
Length = 1124
Score = 47.8 bits (112), Expect = 7e-06
Identities = 32/111 (28%), Positives = 55/111 (48%), Gaps = 1/111 (0%)
Frame = +1
Query: 50 NSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSL 109
+S + SG D+TI ++D+ T + ++ H+ VT++ + NL ++SA DGS
Sbjct: 427 SSYIVSGSFDETIKVWDVKTGKCVHTIKGHTMPVTSVHYNRDGNL-----IISASHDGSC 591
Query: 110 AIFDADGFVHLKTLSVHK-KGVNDLAIHPSGKIALTVSRDSCFAMVNLVRG 159
I+D + LKTL K V+ P+GK+ L + + + N G
Sbjct: 592 KIWDTETGNLLKTLIEDKAPAVSFAKFSPNGKLILAATLNDTLKLWNYGSG 744
Score = 41.6 bits (96), Expect = 5e-04
Identities = 53/249 (21%), Positives = 91/249 (36%)
Frame = +1
Query: 37 PSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPF 96
P+H+ + + ++ S +SGG + L +LTDH + V+ + F + L
Sbjct: 13 PNHV*VSEFRWMATSTTSSGGGGTGTQTF--KPYRHLKTLTDHENAVSCVKFSNDGTL-- 180
Query: 97 PRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNL 156
L SA D +L I+ + L H +G++DLA + S D + +
Sbjct: 181 ---LASASLDKTLIIWSSATLTLCHRLVGHSEGISDLAWSSDSHYICSASDDRTLRIWDA 351
Query: 157 VRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAP 216
G +I+ H +DA +
Sbjct: 352 TVGGGCI-------------------------KILRGH--DDAVFCVNFN---------- 420
Query: 217 AKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASAS 276
++ + +G D I WD+K+GK + I+ H V + D L+ SAS
Sbjct: 421 PQSSYIVSGSFDETIKVWDVKTGKCVHTIK-GHTMPVTSVHYNRDGN-------LIISAS 576
Query: 277 SDGTIRAWD 285
DG+ + WD
Sbjct: 577 HDGSCKIWD 603
>TC211500 similar to UP|Q9FND4 (Q9FND4) WD-repeat protein-like, partial (45%)
Length = 998
Score = 47.4 bits (111), Expect = 9e-06
Identities = 49/241 (20%), Positives = 100/241 (41%), Gaps = 13/241 (5%)
Frame = +2
Query: 57 GSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLV--SADADGSLAIFDA 114
G+ + + L+D+ T + + + V++ +++ P + V S+D + + ++D
Sbjct: 26 GNTEVLKLWDVETGTCKHTFGNQGFVVSSCAWF-----PNSKQFVCDSSDPEKGVCMWDC 190
Query: 115 DGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIV 174
DG V + V DLA+ P G+ +++ D ++++ + +E I
Sbjct: 191 DGNVIKSWRGMRMPKVVDLAVTPDGEYLISIFMDKEIRILHMGTYAERV---ISEEHPIT 361
Query: 175 RFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVL-----------CAAPAKNGLLY 223
VSGDS F V+ +N + + + P R + C N +
Sbjct: 362 SLSVSGDSKFFIVN--LNSQEIHMWDVAGKWDKPLRFMGHKQHKYVIRSCFGGLNNTFIA 535
Query: 224 TGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRA 283
+G E+ + W+ ++ + + H+ V + + P ++ASAS D TIR
Sbjct: 536 SGSENSQVYIWNCRNSRPVEVL-SGHSMTVNCV------SWNPKIPQMLASASDDYTIRI 694
Query: 284 W 284
W
Sbjct: 695 W 697
>TC211342 similar to UP|Q9LVF2 (Q9LVF2) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MIL23, partial (29%)
Length = 862
Score = 44.3 bits (103), Expect = 8e-05
Identities = 46/186 (24%), Positives = 77/186 (40%), Gaps = 22/186 (11%)
Frame = +1
Query: 122 TLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLV--------RGRRSFCCRLDKEASI 173
TL+ HK V L + +G + + SRD+ + ++V RG R + +++
Sbjct: 25 TLNGHKGAVTTLRYNKAGSLLASGSRDNDVILWDVVGETGLFRLRGHRDQAAKQLTVSNV 204
Query: 174 VRFDVSGDSFFMAV------------DEIVNVHQAEDARLLMELQCPKR-VLCAAPAKNG 220
++ D+ +A+ D V VH A+ + + L K VLC + +G
Sbjct: 205 STMKMNDDALVVAISPDAKYIAVALLDSTVKVHFADTFKFFLSLYGHKLPVLCMDISSDG 384
Query: 221 -LLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDG 279
L+ TG D+NI W L G I AHA V + + + + V S D
Sbjct: 385 DLIVTGSADKNIKIWGLDFGDCHKSI-FAHADSVMAVQFV-------PKTHYVFSVGKDR 540
Query: 280 TIRAWD 285
++ WD
Sbjct: 541 LVKYWD 558
Score = 31.6 bits (70), Expect = 0.51
Identities = 57/250 (22%), Positives = 91/250 (35%), Gaps = 13/250 (5%)
Frame = +1
Query: 49 SNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSS------TVTALSFYSPPN-------LP 95
+ S++ASG D+ + L+D+ + L L H TV+ +S + P
Sbjct: 73 AGSLLASGSRDNDVILWDVVGETGLFRLRGHRDQAAKQLTVSNVSTMKMNDDALVVAISP 252
Query: 96 FPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVN 155
+ + A D ++ + AD F +L HK V + I G + +T S D +
Sbjct: 253 DAKYIAVALLDSTVKVHFADTFKFFLSLYGHKLPVLCMDISSDGDLIVTGSADKNIKIWG 432
Query: 156 LVRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAA 215
L G D SI DS MAV + H
Sbjct: 433 LDFG--------DCHKSIF---AHADSV-MAVQFVPKTHY-------------------- 516
Query: 216 PAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASA 275
+++ G+DR + WD ++ +E HA I L+ GD + +
Sbjct: 517 ------VFSVGKDRLVKYWDADKFELLLTLEGHHA----DIWCLAVSNRGD----FIVTG 654
Query: 276 SSDGTIRAWD 285
S D +IR WD
Sbjct: 655 SHDRSIRRWD 684
>TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory protein tup1
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (59%)
Length = 580
Score = 43.9 bits (102), Expect = 1e-04
Identities = 31/112 (27%), Positives = 54/112 (47%), Gaps = 1/112 (0%)
Frame = +3
Query: 50 NSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSL 109
++++ SG D+T+ ++D+ + L L HS VTA+ F +L +VS+ DG
Sbjct: 90 SNIIVSGSFDETVRVWDVKSGKCLKVLPAHSDPVTAVDFNRDGSL-----IVSSSYDGLC 254
Query: 110 AIFDADGFVHLKTL-SVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGR 160
I+DA +KTL V+ + P+ K L + D+ + N G+
Sbjct: 255 RIWDASTGHCMKTLIDDDNPPVSFVKFSPNAKFILVGTLDNTLRLWNYSTGK 410
Score = 34.7 bits (78), Expect = 0.060
Identities = 21/71 (29%), Positives = 33/71 (45%), Gaps = 3/71 (4%)
Frame = +3
Query: 218 KNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDD---EPYLVAS 274
++ ++ +G D + WD+KSGK +K + SD T D + L+ S
Sbjct: 87 QSNIIVSGSFDETVRVWDVKSGKC-----------LKVLPAHSDPVTAVDFNRDGSLIVS 233
Query: 275 ASSDGTIRAWD 285
+S DG R WD
Sbjct: 234 SSYDGLCRIWD 266
>TC203333 similar to UP|Q8H6S5 (Q8H6S5) CTV.2, partial (54%)
Length = 2258
Score = 40.8 bits (94), Expect = 8e-04
Identities = 23/95 (24%), Positives = 45/95 (47%)
Frame = +1
Query: 28 QTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALS 87
+T+T P + + N+++A G D +I +Y++ L H+ +T L+
Sbjct: 1201 KTMTTFMPPPPAATFLAFHPQDNNIIAIGMDDSSIQIYNVRVDEVKSKLKGHTKRITGLA 1380
Query: 88 FYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKT 122
F N+ LVS+ AD + +++ DG+ K+
Sbjct: 1381 FSHVLNV-----LVSSGADAQICVWNTDGWEKQKS 1470
>TC204561 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1419
Score = 40.8 bits (94), Expect = 8e-04
Identities = 52/244 (21%), Positives = 98/244 (39%), Gaps = 4/244 (1%)
Frame = +1
Query: 47 AVSNSVVASGGSDDTIHLYDL--STSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSAD 104
++ N + S D TI L++ ++ HS V+ + F SP L +VSA
Sbjct: 484 SIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDAHSDWVSCVRF-SPSTLQ--PTIVSAS 654
Query: 105 ADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFC 164
D ++ +++ TL+ H VN +A+ P G + + +D + +L G+R +
Sbjct: 655 WDRTVKVWNLTNCKLRNTLAGHNGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLY- 831
Query: 165 CRLDKEASIVRFDVSGDSFFM--AVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLL 222
LD + I S + +++ A ++ + + E ++ +L+ + A
Sbjct: 832 -SLDAGSIIHALCFSPNRYWLCAATEQSIKIWDLESKSIVEDLKVDLKTEADAT------ 990
Query: 223 YTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIR 282
+GG + N KV YC L+ A G + S +DG +R
Sbjct: 991 -SGGGNAN-------KKKVIYCTS------------LNWSADGS----TLFSGYTDGVVR 1098
Query: 283 AWDV 286
W +
Sbjct: 1099VWAI 1110
>TC203656 similar to UP|Q9SYX2 (Q9SYX2) Phytochrome A supressor spa1, partial
(33%)
Length = 1951
Score = 40.8 bits (94), Expect = 8e-04
Identities = 50/248 (20%), Positives = 87/248 (34%), Gaps = 9/248 (3%)
Frame = +2
Query: 53 VASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPP-----NLPFPRNLVSADADG 107
+A+GG I ++DL+ +S D V +S S N +L S D DG
Sbjct: 488 IAAGGVSKKIKIFDLNAIAS--DSVDIQYPVIEMSNKSKLSCVCWNTYIKNHLASTDYDG 661
Query: 108 SLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRL 167
++ ++DAD L H+K
Sbjct: 662 AVQMWDADTGQPLSQYMEHQK--------------------------------------- 724
Query: 168 DKEASIVRFDVSGDSFFMAVDE--IVNVHQAEDARLLMELQCPKRVLCA--APAKNGLLY 223
A V F +S F + + V + + L + P + C + LL+
Sbjct: 725 --RAWSVHFSLSDPKMFASGSDDCSVKLWSISERNSLGTIWKPANICCVQFSAYSTNLLF 898
Query: 224 TGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRA 283
G D + +DL+ ++ +C H V + + EA V SAS+D +++
Sbjct: 899 FGSADYKVYGYDLRHTRIPWCTLAGHGKAVSYVKFIDSEA--------VVSASTDNSLKL 1054
Query: 284 WDVRMAAT 291
WD+ ++
Sbjct: 1055WDLNKTSS 1078
>TC205059 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1492
Score = 39.7 bits (91), Expect = 0.002
Identities = 36/164 (21%), Positives = 74/164 (44%), Gaps = 4/164 (2%)
Frame = +2
Query: 47 AVSNSVVASGGSDDTIHLYDL--STSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSAD 104
++ N + S D TI L++ ++ HS V+ + F SP L +VSA
Sbjct: 560 SIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDAHSDWVSCVRF-SPSTLQ--PTIVSAS 730
Query: 105 ADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFC 164
D ++ +++ TL+ H VN +A+ P G + + +D + +L G+R +
Sbjct: 731 WDRTVKVWNLTNCKLRNTLAGHNGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLY- 907
Query: 165 CRLDKEASIVRFDVSGDSFFM--AVDEIVNVHQAEDARLLMELQ 206
LD + I S + +++ A ++ + + E ++ +L+
Sbjct: 908 -SLDAGSIIHALCFSPNRYWLCAATEQSIKIWDLESKSIVEDLK 1036
Score = 30.8 bits (68), Expect = 0.87
Identities = 50/227 (22%), Positives = 84/227 (36%), Gaps = 18/227 (7%)
Frame = +2
Query: 38 SHLSLIKTVAV---SNSVVASGGSDDTIHLYDLSTSSSLGS-----LTDHSSTVTALSFY 89
+H ++ +A ++ ++ + D +I L+ L+ LT HS V +
Sbjct: 257 AHTDVVTAIATPIDNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLS 436
Query: 90 SPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDS 149
S +S DG L ++D + H K V +A + ++ SRD
Sbjct: 437 SDGQFA-----LSGSWDGELRLWDLAAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDR 601
Query: 150 CFAMVNLVRGRRSFCCRLDKEA-----SIVRFDVSG---DSFFMAVDEIVNVHQAEDARL 201
+ N + G + + D +A S VRF S + D V V + +L
Sbjct: 602 TIKLWNTL-GECKYTIQ-DGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTNCKL 775
Query: 202 LMELQCPKRVL--CAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIE 246
L + A L +GG+D I WDL GK Y ++
Sbjct: 776 RNTLAGHNGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLYSLD 916
>TC226869 similar to UP|Q8H6S5 (Q8H6S5) CTV.2, partial (40%)
Length = 1800
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/90 (25%), Positives = 41/90 (45%)
Frame = +2
Query: 28 QTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALS 87
+T+T P + + N+++A G D +I +Y++ L H +T L+
Sbjct: 743 KTMTTFMPPPPAATFLAFHPQDNNIIAIGMEDSSIQIYNVRVDEVKTKLKGHQKRITGLA 922
Query: 88 FYSPPNLPFPRNLVSADADGSLAIFDADGF 117
F N+ LVS+ AD L ++ DG+
Sbjct: 923 FSHVLNV-----LVSSGADSQLCVWSTDGW 997
>TC229333 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial (32%)
Length = 982
Score = 38.5 bits (88), Expect = 0.004
Identities = 25/88 (28%), Positives = 44/88 (49%), Gaps = 1/88 (1%)
Frame = +3
Query: 52 VVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAI 111
++ASGG D + L+ + +L +HSS +T + F P++P L ++ D ++ +
Sbjct: 702 LLASGGHDKRVVLWYTDSLKQKATLEEHSSLITDVRF--SPSMP---RLATSSFDKTVXV 866
Query: 112 FDADG-FVHLKTLSVHKKGVNDLAIHPS 138
+D D L T + H V L HP+
Sbjct: 867 WDVDNPGYSLHTFTGHSTSVMSLNFHPN 950
>TC221163
Length = 832
Score = 38.5 bits (88), Expect = 0.004
Identities = 26/89 (29%), Positives = 44/89 (49%), Gaps = 1/89 (1%)
Frame = +3
Query: 51 SVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLA 110
S VASG D + L+D + + +L HS + +LS S N LVSA DG+
Sbjct: 495 SYVASGCVDGKVRLWDSRSGECVKTLKGHSDAIQSLSVSSNRNY-----LVSASVDGTAC 659
Query: 111 IFDADGFV-HLKTLSVHKKGVNDLAIHPS 138
F+ + F + + +H+ ++ + +PS
Sbjct: 660 AFEVENFR*TIPVVLLHRMRLSFIQANPS 746
Score = 32.0 bits (71), Expect = 0.39
Identities = 53/275 (19%), Positives = 95/275 (34%), Gaps = 9/275 (3%)
Frame = +3
Query: 59 DDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFD---AD 115
D +I +++ ++ L + H +VT F P + + + D +L I++ +
Sbjct: 6 DFSIWMWNTDNAALLNTFIGHGDSVTCGDF-----TPDGKIICTGSDDATLRIWNPKTGE 170
Query: 116 GFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVR 175
++ H +G+ L I+ + +AL+ S+D +VN+ GR +D A
Sbjct: 171 STHVVRGHPYHTEGLTCLTINSTSTLALSGSKDGSVHIVNITTGRV-----VDNNALASH 335
Query: 176 FDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGL-LYTGGEDRNITAW 234
D + C A +G GG D+ + W
Sbjct: 336 SD--------------------------------SIECVGFAPSGSWAAVGGMDKKLIIW 419
Query: 235 DLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDVRMAATEK- 293
D++ + + G+ L+ VAS DG +R WD R K
Sbjct: 420 DIE-----HLLPRGTCEHEDGVTCLAWLGAS-----YVASGCVDGKVRLWDSRSGECVKT 569
Query: 294 ----SEPLAECKTQSRLTCLAGSSLKCKLKKFEFE 324
S+ + S L +S+ FE E
Sbjct: 570 LKGHSDAIQSLSVSSNRNYLVSASVDGTACAFEVE 674
>TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%)
Length = 1048
Score = 38.5 bits (88), Expect = 0.004
Identities = 25/82 (30%), Positives = 45/82 (54%), Gaps = 2/82 (2%)
Frame = +2
Query: 36 YPSHLSLIKTVAVS--NSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPN 93
+ H +I ++A+S +ASG D TI ++DLS+ L L H+S V +L+F S +
Sbjct: 353 FVGHRGMILSLAMSPDGRYMASGDEDGTIMMWDLSSGRCLTPLIGHTSCVWSLAFSSEGS 532
Query: 94 LPFPRNLVSADADGSLAIFDAD 115
+ + S AD ++ ++D +
Sbjct: 533 V-----IASGSADCTVKLWDVN 583
Score = 32.3 bits (72), Expect = 0.30
Identities = 21/63 (33%), Positives = 32/63 (50%)
Frame = +2
Query: 224 TGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRA 283
TG D+ + WD++SG+ C+ R G+++ A D Y+ AS DGTI
Sbjct: 290 TGSSDKTVRLWDVQSGE---CVRVFVGHR--GMIL--SLAMSPDGRYM-ASGDEDGTIMM 445
Query: 284 WDV 286
WD+
Sbjct: 446 WDL 454
>TC232143 similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-like, partial (22%)
Length = 608
Score = 38.1 bits (87), Expect = 0.005
Identities = 31/130 (23%), Positives = 60/130 (45%), Gaps = 9/130 (6%)
Frame = +1
Query: 167 LDKEASIVRFDVSGDSFFMAVDEI---VNVHQAE-DARLLMELQCPKRVL-----CAAPA 217
+D++ +I F +S DS + V+ + +++ E D +L+ + + KR C
Sbjct: 19 IDEDQTITSFSLSKDSRLLLVNLLNQEIHLWNIEGDPKLVGKYRSHKRSRFVIRSCFGGL 198
Query: 218 KNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASS 277
+ + +G ED + W SG + + H+ V + + P+++ASAS
Sbjct: 199 EQSFIASGSEDSQVYIWHRSSGDLIETLP-GHSGAVNCV------SWNPANPHMLASASD 357
Query: 278 DGTIRAWDVR 287
D TIR W ++
Sbjct: 358 DRTIRIWGLK 387
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,495,962
Number of Sequences: 63676
Number of extensions: 192817
Number of successful extensions: 1227
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 1139
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1212
length of query: 324
length of database: 12,639,632
effective HSP length: 97
effective length of query: 227
effective length of database: 6,463,060
effective search space: 1467114620
effective search space used: 1467114620
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144656.10


